Evaluation of Synergistic Effect of Predicted Network Pharmacology of Berberine in Type 2 Mellitus Diabetes Treatment
Li Bo, Chen Yu-Bao*, Hu Wen-Xiang, Pan Chen-Ling and Meng Hao
Published Date: 2025-01-21Li Bo1, Chen Yu-Bao2*, Hu Wen-Xiang3, Pan Chen-Ling4 and Meng Hao5
1Department of Chemistry and Environmental Engineering, Wuhan Institute of Technology, Wuhan 430205, China
2Department of Medical Sciences, Chinese Academy of Medical Sciences, Institute of Medical Laboratory Animal Science, Beijing 100021, China
3Department of Medical Sciences, Beijing Shenjian Tianjun Academy of Medical Sciences, Beijing 101601, China
4Department of Computer Science, Beijing Computing Center, Beijing 102209, China
5Department of Medicine Technology, Peking University Medicine Technology, Transfer and Innovation Center, Beijing 100191, China
- *Corresponding Author:
- Chen Yu-Bao
Department of Medical Sciences, Chinese Academy of Medical Sciences, Institute of Medical Laboratory Animal Science, Beijing 100021, China
E-mail:allexchen@126.com
Received date: June 10, 2024, Manuscript No. IPJHM-24-19154; Editor assigned date: June 12, 2024, PreQC No. IPJHM-24-19154 (PQ); Reviewed date: June 26, 2024, QC No. IPJHM-24-19154; Revised date: January 14, 2025, Manuscript No. IPJHM-24-19154 (R); Published date: January 21, 2025, DOI: 10.36648/2472-0151.11.1.003
Citation: Bo L, Yu-Bao C, Wen-Xiang H, Chen-Ling P, Hao M (2025) Evaluation of Synergistic Effect of Predicted Network Pharmacology of Berberine in Type 2 Mellitus Diabetes Treatment. Herb Med Vol:11 No:1
Abstract
Objectives: Based on the compounds-target-disease network theory, network pharmacology are new approaches for systematic studying the diseases, revealing the mystery of active compounds synergistic effect and investigating the mechanism of Traditional Chinese Medicines (TCM). The potential pharmacodynamic material basis and network regulation mechanism of berberine in diabetes and related chronic diseases are studied.
Methods: The corresponding ID number of berberine was checked in the PubChem database. The putative targets of diabetes and related chronic diseases were found through the IPA commercial database combined with OMIM database. Finally, it established a berberine-target genesdiseases network by Cytoscape. Graph-based techniques are examined by the network diffusion methods to predict berberine-target genes-diseases network and influence the transmission in networks.
Results: The corresponding gene of berberine were found in the IPA database, and the effective targets for diabetes and related chronic diseases were obtained by combining OMIM. It is confirmed that berberine worked on 45 putative targets, such as APOA1, VEGFA, PPARG, PRKCZ, TNF etc. These drug-target networks involve about 20 diseases such as diabetes, high blood pressure, coronary heart disease, cardiac arrhythmia, depression, neuro division, and so on. On the basis of above similarities and interaction data for gene and targets; a new gene or target get its interaction profile respectively; a new gene is one that has unknown targets and in the similar manner a new target is the one which has no known interactions with any genes and proposed efficient way to understand berberine pathways mechanisms.
Conclusion: The common putative targets of diabetes and related chronic diseases are identified from the molecular level, established the berberine-drug target predictiondisease pharmacology network. This method may offer an efficient way to understand the berberine, multi-targets and multiple pathways mechanisms of berberine for treatment of diabetes and related chronic diseases.
Keywords
Berberine; Network pharmacology; Diabetes; Chronic diseases
Introduction
Berberine is the main active ingredient of Coptidis. It can be extracted from traditional Chinese medicines such as Coptis, Phellodendron and Berberis sargentiana. It is an alkaloid and is often used as an anti-infective and anti-inflammatory drug for bacterial gastroenteritis. It is also used for gastrointestinal tract such as dysentery and other digestive diseases. Recent studies have shown that berberine also plays an important role in the treatment of medical diseases, oncological diseases, and antidiabetics and antihyperlipidemic [1-4]. The therapeutic effect of berberine is mainly manifested in its pharmacological effects such as blood sugar lowering, blood lipids, antiinflammatory and anti-oxidation, and it has well relief and protection for diabetes complications such as diabetes and diabetic nephropathy, diabetic cardiovascular disease and diabetic peripheral neuropathy. Based on the therapeutic effect of berberine, this study mainly analyzes the numerous networkcoordinated therapeutic effects of berberine, focusing on the network mechanism of berberine in the treatment of diabetes and other chronic diseases.
Diabetes is a group of metabolic diseases characterized by hyperglycemia and characterized by disorders of glucose metabolism. It is caused by defects in insulin secretion and its biological effects. Hyperglycemia can cause lesions in various tissues of the body, especially chronic damage and dysfunction of the eyes, kidneys, heart, and nerves. In recent years, the incidence of diabetes has continued to rise worldwide, and the World Health Organization (WHO) international diabetes federation survey shows that there are nearly 4 billion people with diabetes worldwide [5,6]. Diabetes has become the third non-infectious chronic disease that threatens human health after cardiovascular disease and cancer on a global scale, and the number of diabetic patients in China is increasing [7].
Diabetes belongs to the category of Chinese medicine, and it has a long history of using traditional Chinese medicine to treat diabetes. Many studies have also shown that Chinese medicine also plays an important role in the treatment of diabetes. The study of berberine (also known as berberine) in traditional Chinese medicine for the treatment of diabetes has received increasing attention [8]. It has also been found that berberine can reduce the levels of fasting blood glucose, postprandial blood glucose, glycosylated hemoglobin and triglycerides in patients with type 2 diabetes, and its hypoglycemic and lipidlowering effects may be multi-target. At present, there are more studies on berberine molecular, but the mechanism of action is not very clear. In this paper, the network pharmacology method was used to establish the berberine-multi-gene-multi-disease network model by means of the Ingenuity Pathway Analysis (IPA) commercial database and the Online Mendelian Inheritance in Man (OMIM) open-source database to elucidate the mechanism of action of the main components of berberine. It lays a further molecular theoretical foundation for the treatment of diabetes and other chronic diseases by berberine, multi-target, and multipath.
Materials and Methods
Determination of berberine and diabetes related genes
This study strives to accurately determine the mechanism of action of berberine molecules in the treatment of diabetes and other chronic diseases at the genetic level. Therefore, this study first determined the corresponding relationship between berberine molecule and the corresponding genes of diabetes and other chronic diseases. The specific implementation process is as follows: The CID number of berberine molecule is imported into the Ingenuity Pathway Analysis (IPA) commercial database, and the search range is determined at the time of inquiry. The selected species, the direct or indirect interactions were determined, and the target gene information corresponding to the introduced berberine molecule was finally retrieved. Table 1 shows the genes corresponding to berberine inquired by the Ingenuity Pathway Analysis (IPA) commercial database. Then look for all other genetic information for diabetes and other chronic diseases in the human Mendelian genetic database, the Online Mendelian Inheritance in Man (OMIM) open-source database. Finally, the genes searched by the two databases are matched by the gene ID, and the genetic information of the two databases is extracted by the common gene ID number to extract the common gene obtained from the two databases, so that the chemical component-gene-correspondence of disease can be obtained. Related studies indicate that there are 15 susceptibility genes for type 2 diabetes in Chinese population: ADIPOQ, CDH13, CDNK2B, GLIS3, GRB14, HHEX, HNF1A, IRS1, KCNQ1, PAX4, PEPD, PPARG, RASGRP1, SLC30A8, TCF7L2. The query found that three genes in Table 1 are type 2 diabetes susceptibility genes in the Chinese population, namely ADIPOQ, PPARG and IRS1.
| Berberine | ADIPOQ | SCARB1 | NOS2 | IL10 | ERBB2 | CDK2 |
| Berberine | APOA1 | STAT1 | PCSK9 | IL6 | FOXO1 | CDK4 |
| Berberine | APOB | STAT3 | PON1 | INSR | GCG | CYPIA2 |
| Berberine | AR | TGFB1 | PPARA | IRS1 | GRK2 | CYP2E1 |
| Berberine | BIRC5 | TH | PPARG | LDLR | HIF1A | CYP3A5 |
| Berberine | CASP1 | TNF | PRKAA1 | MAPT | HMGB1 | CYP4A11 |
| Berberine | CASP3 | TP53 | PRKCZ | MMP2 | HMGCR | ENDOG |
| Berberine | CCL2 | VEGFA | PTGER2 | MMP9 | HRAS |
Table 1: Related genes of berberine.
Construct berberine-gene-disease correlation
In this paper, the network model of berberine-multi-genemultiple diseases was established by the professional network analysis software which named Cytoscape. The network model includes three network nodes which are the active components of berberine, related target genes and diabetes and other chronic diseases. The connection between nodes and nodes indicates that there is interaction between the two nodes, so the three kinds of nodes have a total of five correspondences, namely berberine active ingredient-target gene-diseases, target gene-berberine active ingredient-target genes, diseases-target genes-diseases, diseases-target genes-berberine active ingredients, target genes-diseases-targets gene. The network model of berberine-multi-gene-multiple diseases was established through the above five corresponding interactions, and then the mechanism of action of berberine and corresponding diseases could have analyzed.
Network diffusion models: Graph-based techniques are examined by the network diffusion methods to predict berberine-target genes-diseases network and influence the transmission in networks. To train a network can well classify, we require a list of previously known berberine-target genesdiseases network. In other way we want to predict the new network interaction which is not based on existing training data. The data which represents the gene and the target involved in network is also needed for this purpose. Neighborhood methods perform predictions on the basis of functions that are relatively simple. More accurately, on the basis of previous similarities and interaction data for gene and targets, a new gene or target can get its interaction profile respectively, a new gene is one that has unknown targets and in the similar manner a new target is the one which has no known interactions with any genes.
Result
Analysis of berberine related gene
The genes corresponding to berberine found in the Ingenuity Pathway Analysis (IPA) commercial database are shown in Table 1. We can see that the number of target genes corresponding to berberine is 45, that is, berberine can regulate the expression of 45 genes such as APOA1, VEGFA, PPARG, PRKCZ, TNF, and so on. Berberine have different prevention and treatment effects on the diseases involved in these genes. There are three diabetes susceptibility genes are susceptibility genes associated with type 2 diabetes in the Chinese population, which are the ADIPOQ gene, the PPARG gene, and the IRS1 gene, respectively.
In addition to the direct relationship between the ADIPOQ gene and type 2 diabetes in the Chinese population, the protein encoded by this gene is also closely related to the formation of fatty liver. The PPARG gene is a peroxisome proliferatoractivated receptor gamma-encoding gene, which belongs to the nuclear receptor peroxisome proliferator-activated receptor subfamily. It can bind to the retinoid receptor to form a heterodimer, and the binding of DAN regulates the transcription of a variety of genes. PPARG is mainly responsible for the catabolism of lipids, and is also associated with a variety of diseases, including obesity, diabetes, arteriosclerosis and cancer. PPARG is expressed in different degrees in tissues such as fat, liver, skeletal muscle, kidney, and pancreas. It is activated by binding to ligands, and activates or inhibits genes by interacting with PPAR Response Elements (PPREs) in the promoter of the target gene. The transcription of thus participate in a variety of physiological and pathological processes in the body, including adipocyte differentiation, inflammatory response, apoptosis, obesity, atherosclerosis and cancer. PPARG can be activated by fatty acids and plays an important role in insulin sensitivity and fat production. An increase in PPARG activity increases the absorption of fatty acids and the accumulation of fat, which in turn produces obesity. IRS1 is the major substrate for insulin receptor kinase. IRS1 contains multiple sites of tyrosine phosphorylation that phosphorylate to bind to proteins containing the SH domain, thereby mediating insulin-induced metabolism and growthpromoting signals. IRS1 mediates insulin signaling on the one hand, but IRS1 overexpression or some serine phosphorylation also leads to insulin resistance.
Statistical analysis of diabetes related gene
The genes associated with diabetes and other chronic diseases were searched by the Online Mendelian Inheritance in Man (OMIM) open source database. The genes found are shown in Table 2. It can be seen that the number of genes associated with different diseases is also different, wherein the number of genes associated with hypertension is 23, the number of genes associated with diabetes is 13, and the number of genes associated with depression is 6, the number of genes associated with arrhythmia is 7 and so on. The chronic diseases mentioned in this study mainly include chronic diseases such as hypertension, depression, coronary heart disease, and arrhythmia. Among the genetic information in Table 1, three genes (ADIPOQ gene, PPARG gene and IRS1 gene) are directly related to type 2 diabetes in the Chinese population. Table 2 shows that the number of genes associated with diabetes is 13. The data in Table 2 is directly from the disease, directly query the genetic information related to it, here contains the genetic information directly related and indirectly related. At the same time, because there are multiple genes may be related to a variety of diseases, such as PPARG gene in addition to lipid catabolism, but also related to a variety of diseases, including obesity, diabetes, adipocyte differentiation, inflammation, apoptosis, arteriosclerosis and cancer.
| Disease | Number of gene | Disease | Number of gene | Disease | Number of gene |
| Anxiety | 1 | Depression | 4 | Tension | 1 |
| Blues | 1 | Depressive | 1 | The neuroses syndrome | 2 |
| Care | 3 | Diabetes | 13 | Heart failure | 7 |
| Chest distress | 2 | Dyspnea | 2 | High blood pressure | 15 |
| Concern | 3 | Eacute myocardial infarction | 11 | Insomnia | 1 |
| Hypertension | 8 | Fear | 1 | Neuro division | 6 |
| Depressed | 1 | Palpitation | 1 |
Table 2: Diabetes and other chronic diseases correspond to other genes.
Berberine-multi-gene-multiple disease network analysis
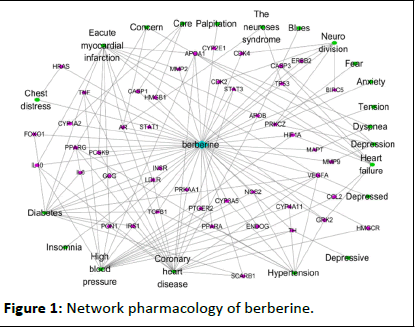
In order to study the regulation of berberine on diabetes and other chronic diseases, berberine was subjected to componentmulti- gene-multiple disease analysis. There are many gene targets that can be regulated by berberine from IPA database. We integrate with the genes corresponding to the diseases queried by OMIM, and finally use Cytoscape software to map out the main component of berberine-multi-gene-multi-disease network. The model is shown in Figure 1. In the figure, the innermost layer is represented by a light blue diamond-shaped node, which is a berberine molecule; the purple rectangular node represents the target gene information that can be regulated by berberine; and the green elliptical node represents other disease information corresponding to the target gene. The components, genes, and diseases are not layered in the figure, and are intended to show other diseases of gene regulation clearly. It can be clearly seen from the figure that berberine mainly regulates the expression of 45 genes such as APOA1, VEGFA, PPARG, PRKCZ, TNF, etc., which associated with and diabetes and other chronic diseases. The main diseases are diabetes, hypertension, coronary heart disease, disorders, depression, schizophrenia, etc. Among them, the APOA1 gene encodes serum apolipoprotein A1, which is a protective factor for atherosclerosis. Its content and contrast value are important for the prevention and judgment of atherosclerosis and cardiovascular disease. The full name of VEGFA is vascular endothelial growth factor A, which is widely distributed in many tissues of the human body, such as gland, liver, lung, kidney, heart machine, etc., which plays a key role in tumor angiogenesis and invades tumors. PPARG gene is involved in a variety of diseases. PPARG gene is responsible for lipid catabolism, and chronic diseases related to obesity, diabetes, adipocyte differentiation, inflammatory response, apoptosis, arteriosclerosis and cancer.
Figure 1: Network pharmacology of berberine.
Molecular docking
Due to the complex diversity of its constituents, traditional Chinese medicines often have a complicated mechanism of action in treating diseases. According to the theory of receptors, the chemical composition of traditional Chinese medicine is still the material basis of biological activity, and its role is to interact with other active functional receptors. Therefore, computer and other professional software can be used to simulate the direct interaction between components and target molecules. This has become a common research method in the field of drug research, and is gradually being applied to the research of traditional Chinese medicine [9-16].
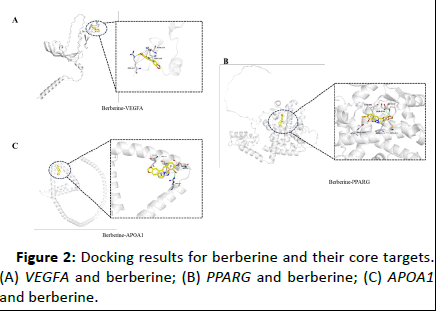
Therefore, the berberine can regulate more genes and some of them for partial molecular docking verification. Here the berberine molecule and serum apolipoprotein A1, vascular endothelial growth factor A protein, peroxidation which encoding by APOA1, VEGFA, PPARG genes performed molecular docking on the interaction between the receptor and the protein target. In this paper, the Autodock Vina program is used for semi-flexible molecular docking calculations, that is, only the conformation of the berberine molecule is changed during the docking process, and the structure of the receptor protein remains unchanged. The position of the lattice box of the receptor binding site in the docking process is set in the position reported in the literature, and finally the optimal binding conformation is found based on the semi-empirical free energy function. Through molecular docking, it was found that the berberine molecule has a strong interaction with the above three target proteins, docking scores are shown in Table 3, docking results for berberine and their core targets shown in Analysis (IPA) database and the Online Mendelian Inheritance in Man (OMIM) database to provide a network model for the regulation of diabetes and other chronic diseases by berberine, which has certain guiding significance for subsequent scientific research.
| Ligands | Receptors | ||
| APOA1 | VEGFA | PPARG | |
| Berberine | -7.2 | -6.6 | -9.4 |
Table 3: Molecular docking scores (kcal/mol).
Figure 2: Docking results for berberine and their core targets. (A) VEGFA and berberine; (B) PPARG and berberine; (C) APOA1 and berberine.
KEGG enrichment analysis
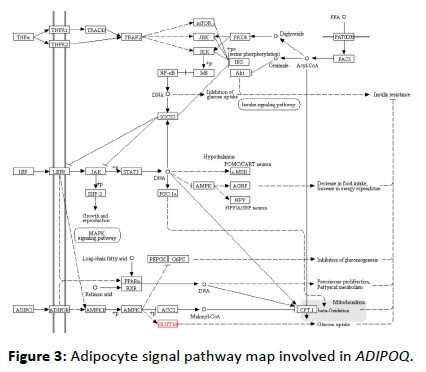
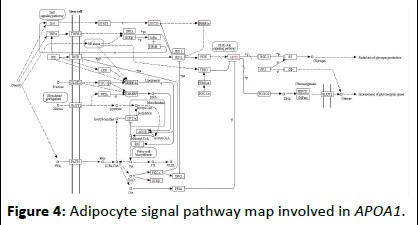
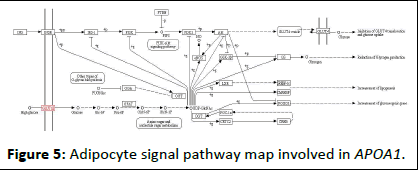
The above molecular docking technique verified the interaction between the berberine molecule and the target protein molecules encoded by the three genes APOA1, VEGFA and PPARG, and proved that there is a strong interaction between between them. For the network mechanism of berberine, we performed KEGG pathway enrichment analysis on all target genes corresponding to berberine. The pathway enrichment map is shown in Table 4. The pathway enrichment map shows the pathways involved in the 45 genes of the berberine component corresponding to the target. In each pathway, the target gene realizes its regulatory function through its upstream and downstream genes, thus completing its intricate pathway to disease treatment. In this study, an APOA1 gene closely related to adipokines and diabetes was selected, and its KEGG pathway related to diabetes was analyzed, as shown in Figures 3-5. For each pathway, it contains extremely complicated upstream and downstream regulation relationships. Therefore, we only give the enrichment analysis of the KEGG pathway. The corresponding regulation process of each pathway will be given one by one in future research. At the same time, this study gave the distribution of the biological pathways of type 2 diabetes susceptibility genes in Chinese population corresponding to berberine, as shown in Table 5.
| Category | Term | Genes | Benjamini |
| KEGG_PATHWAY | Pathways in cancer | PTGER2, IL6, MMP9, ERBB2, PPARG, TP53, FOXO1, BIRCS, STATI, CDK4, MMP2, TGFB1, STAT3, CDK2, CASP3, HIFIA, VEGFA, N | 4.30E-11 |
| KEGG_PATHWAY | Hepatitis B | CASP3, IL6, TNF, MMP9, TP53, BIRC5, CDK4, STAT1, TGFB1, CDK2, STAT3 | 4.40E-07 |
| KEGG_PATHWAY | Insulin resistance | PPARA, PRKCZ, IL6, TNF, FOXO1, PRKAA1, INSR, IRS1, STAT3 | 6.60E-06 |
| KEGG_PATHWAY | Non-alcoholic fatty liver disease | PPARA, CASP3, IL6, TNF, PRKAA1, CYP2E1, ADIPOQ, INSR, IRSI, TGFBI | 5.SE-06 |
| KEGG_PATHWAY | FoxO signaling pathway | IL6, FOXO1, PRKAAI, INSR, IRSI, TGFB1, CDK2, IL10, STAT3 | 1.50E-05 |
| KEGG PATHWAY | Proteoglycans in cancer | CASP3, HIFIA, TNF, MMP9, ERBB2, VEGFA, TP53, MMP2, TGFB1, STAT3 | 1.70E-05 |
| KEGG_PATHWAY | Pancreatic cancer | ERBB2, VEGFA, TP53, CDK4, STAT1, TGFB1, STAT3 | 2.20E-05 |
| KEGG_PATHWAY | Toxoplasmosis | CASP3, TNF, LDLR, NOS2, STAT1, TGFB1, IL10, STAT3 | 2.60E-05 |
| KEGG_PATHWAY | Bladder cancer | MMP9, ERBB2, VEGFA, TP53, CDK4, MMP2 | 2.90E-05 |
| KEGG_PATHWAY | HIF-1 signaling pathway | IL6, HIFIA, ERBB2, VEGFA, NOS2, INSR, STAT3 | 2.00E-04 |
| KEGG_PATHWAY | Inflammatory Bowel Disease (IBD) | IL6, TNF, STAT1, TGFB1, IL10, STAT3 | 2.60E-04 |
| KEGG_PATHWAY | AMPK signaling pathway | HMGCR, PPARG, FOXO1, PRKAA1, ADIPOQ, INSR, IRSI | 4.80E-04 |
| KEGG_PATHWAY | Adipocytokine signaling pathway | PPARA, TNF, PRKAAI, ADIPOQ, IRSI, STAT3 | 4.60E-04 |
| KEGG_PATHWAY | Pertussis | CASP3, IL6, TNF, NOS2, CASP1, IL10 | 4.90E-04 |
| KEGG_PATHWAY | Hepatitis C | PPARA, TNF, LDLR, TP53, SCARB1, STAT1, STAT3 | 6.20E-04 |
| KEGG_PATHWAY | Type II diabetes mellitus | PRKCZ, TNF, ADIPOQ, INSR, IRSI | 1.30E-03 |
| KEGG_PATHWAY | Malaria | IL6, CCL2, TNF, TGFB1, IL10 | 1.60E-03 |
| KEGG_PATHWAY | Amoebiasis | CASP3, IL6, TNF, NOS2, TGFB1, IL10 | 2.10E-03 |
| KEGG_PATHWAY | Chagas disease (American trypanosomiasis) | IL6, CCL2, TNF, NOS2, TGFB1, IL10 | 2.20E-03 |
| KEGG_PATHWAY | Tuberculosis | CASP3, IL6, TNF, NOS2, STAT1, TGFB1, IL10 | 2.60E-03 |
| KEGG_PATHWAY | Leishmaniasis | TNF, NOS2, STAT1, TGFB1, IL10 | 2.50E-03 |
| KEGG_PATHWAY | Herpes simplex infection | CASP3, IL6, CCL2, TNF, TP53, STATI, CDK2 | 3.00E-03 |
| KEGG_PATHWAY | Measles | IL6, TP53, CDK4, STATI, CDK2, STAT3 | 4.SE-03 |
| KEGG_PATHWAY | African trypanosomiasis | IL6, APOA1, TNF, IL10 | 5.20E-03 |
| KEGG_PATHWAY | Prostate cancer | AR, ERBB2, TP53, FOXO1, CDK2 | 7.00E-03 |
| KEGG_PATHWAY | Rheumatoid arthritis | IL6, CCL2, TNF, VEGFA, TGFBI | 7.00E-03 |
| KEGG PATHWAY | MicroRNAs in cancer | CASP3, MMP9, ERBB2, VEGFA, TP53, IRSI, STAT3 | 8.00E-03 |
| KEGG_PATHWAY | PISK-Akt signaling pathway | IL6, VEGFA, TP53, PRKAA1, CDK4, INSR, IRS1, CDK2 | 9.SE-03 |
| KEGG_PATHWAY | NOD-like receptor signaling pathway | IL6, CCL2, TNF, CASP1 | 1.20E-02 |
| KEGG_PATHWAY | TNF signaling pathway | CASP3, IL6, CCL2, TNF, MMP9 | 1.40E-02 |
| KEGG_PATHWAY | Amyotrophic Lateral Sclerosis (ALS) | CASP3, TNF, TP53, CASPI | 1.60E-02 |
| KEGG_PATHWAY | Legionellosis | CASP3, IL6, TNF, CASP1 | 2.20E-02 |
| KEGG_PATHWAY | Insulin signaling pathway | PRKCZ, FOXO1, PRKAA1, INSR, IRSI | 2.50E-02 |
| KEGG_PATHWAY | PPAR signaling pathway | PPARA, APOA1, PPARG, ADIPOQ | 2.50E-02 |
| KEGG PATHWAY | Colorectal cancer | CASP3, TP53, BIRCS, TGFB1 | 2.60E+00 |
| KEGG_PATHWAY | Apoptosis | CASP3, TNF, ENDOG, TP53 | 2.60E-02 |
| KEGG_PATHWAY | p53 signaling pathway | CASP3, TP53, CDK4, CDK2 | 2.90E-02 |
| KEGG_PATHWAY | Hypertrophic Cardiomyopathy (HCM) | IL6, TNF, PRKAAI, TGFBI | 3.50E-02 |
| KEGG PATHWAY | Transcriptional misregulation in cancer | IL6, MMP9, PPARG, TP53, FOXO1 | 3.60E-02 |
| KEGG_PATHWAY | Small cell lung cancer | TP53, NOS2, CDK4, CDK2 | 4.10E-02 |
| KEGG_PATHWAY | Influenza A | IL6, CCL2, TNF, STAT1, CASP1 | 4.70E-02 |
Table 4: KEGG pathway enrichment table of berberine corresponding to target.
| Gene | Pathway |
| ADIPOQ | Non-Alcoholic Fatty Liver Disease (NAFLD); AMPK signaling pathway; PPAR signaling pathway; Type II diabetes mellitus |
| PPARG | Pathways in cancer; AMPK signaling pathway; PPAR signaling pathway; Transcriptional misregulation in cancer |
| IRS1 | Insulin resistance; Non-Alcoholic Fatty Liver Disease (NAFLD); FoxO signaling pathway; MicroRNAs in cancer; PI3K-Akt signaling pathway; Insulin signaling pathway |
Table 5: Distribution of susceptibility gene biological pathways in type 2 diabetes in Chinese population.
Figure 3: Adipocyte signal pathway map involved in ADIPOQ.
Figure 4: Adipocyte signal pathway map involved in APOA1.
Figure 5: Adipocyte signal pathway map involved in APOA1.
Network visualization
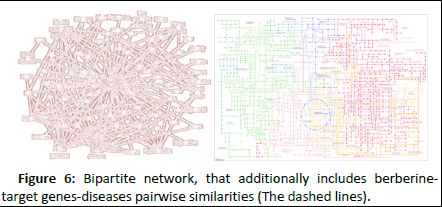
Network visualization as an experimental study aid, may be used to demonstrate the berberine-target genes-diseases bipartite network. Examining the network visually may provide some evidences or understandings which may be difficult to reach otherwise. For example, by examining closely the visualized network carefully for hints, it is possible to tell why certain interactions have low prediction score. The user may use the edge width or coloring edges with a color scale to specify how high or low the prediction scores may be. This particular example is demonstrated in Figure 6.
Network diffusion model
The methods which involves investigation of graph-based techniques to predict new interactions falls in the network diffusion category. It is named as such because it is predominant in this category. Network-Based Inference (NBI) applies network diffusion on the berberine-target genes-diseases bipartite network corresponding to the interaction matrix Y to perform predictions. The working of network diffusion is according to:Y=WY
Where W ∈Rn×n is the weight matrix defined as:
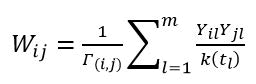
Where Γ is the diffusion rule, and k(x) denotes the degree of node x in the berberine-target genes-diseases bipartite network. While in NBI case, the Γ rule is given by Γ=k(dj).
Figure 6: Bipartite network, that additionally includes berberinetarget genes-diseases pairwise similarities (The dashed lines).
Discussion
In this paper, the network pharmacology research method was used to analyze the network model of berberine regulating diabetes and other chronic diseases, and the molecular docking method was used to verify the mechanism. It was found that berberine can regulate the expression APOA1, VEGFA, PPARG, PRKCZ, TNF, etc. which involves diabetes, hypertension, coronary heart disease, arrhythmia, depression, schizophrenia and other diseases.
Conclusion
In summary, this paper uses the computational method to study the network model of berberine-multi-gene-multiple diseases, avoiding the barriers that are difficult to obtain in the chemical composition of traditional Chinese medicine, and provides calculation hypothesis and experimental direction for further research. This method reduce the amount of experimental work, which in turn reduces the cost of new drug development and research in the future. Therefore, the use of network to study the role of TCM in treating diseases and preventing disease has given new scientific connotations to traditional Chinese medicine. At the same time, this result has a good reference for the study of complex diseases based on metabolic pathways.
Acknowledgement
Thanks to Professor Wei Dongqing for his guidance.
Competing and Conflicting Interests
The authors declare no competing financial interest.
References
- Song J (2008) New progress in clinical application and research of Xiaochai. Mod Med Health 24:1652-1655
- Li H, Chai X, Li X F (2008) Clinical use and adverse reactions of berberine. Armed Police Med 19:661-662
- Pang B, Zhao LH, Zhou Q, Zhao TY, Wang H, et al. (2015) Application of berberine on treating type 2 diabetes mellitus. Int J Endocrinol 2015:905749
[Crossref] [Google Scholar] [PubMed]
- Liu C, Wang Z, Song Y, Wu D, Zheng X, et al. (2015) Effects of berberine on amelioration of hyperglycemia and oxidative stress in high glucose and high fat dietâ?ÂÂÂÂÃÂinduced diabetic hamsters in vivo. Biomed Res Int 2015:313808
[Crossref] [Google Scholar] [PubMed]
- Tai N, Wong FS, Wen L (2015) The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev Endocr Metab Disord 16:55-65
[Crossref] [Google Scholar] [PubMed]
- El-Kaissi S, Sherbeeni S (2011) Pharmacological management of type 2 diabetes mellitus: An update. Curr Diabetes Rev 7:392-405
[Crossref] [Google Scholar] [PubMed]
- Fan LY, Du JL (2012) Diabetes hazards and prevention. Pract J Intern Med 26:56-59
- Yin J, Xing H, Ye J (2008) Efficacy of berberine in patients with type 2 diabetes mellitus. Metabolism 57:712-717
[Crossref] [Google Scholar] [PubMed]
- Hood L, Heath JR, Phelps ME, Lin B (2004) Systems biology and new technologies enable predictive and preventative medicine. Science 306(5696):640-643
[Crossref] [Google Scholar] [PubMed]
- di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L (2006) From in vivo to in silico biology and back. Nature 443(7111):527-533
[Crossref] [Google Scholar] [PubMed]
- Ekins S (2004) Predicting undesirable drug interactions with promiscuous proteins in silico. Drug Discov Today 9:276-285
[Crossref] [Google Scholar] [PubMed]
- Wu DH, Xu XJ (2009) Computer simulation of the mechanism of qi and blood circulation of traditional Chinese medicine. J Phys Chem 25:446-450
- Huang Q, Qiao X, Xu X (2007) Potential synergism and inhibitors to multiple target enzymes of Xuefu Zhuyu Decoction in cardiac disease therapeutics: A computational approach. Bioorg Med Chem Lett Letters 17:1779-1783
[Crossref] [Google Scholar] [PubMed]
- Qin H, Xuebing Q, Xiaojie X (2007) Chemical analysis of effective components of Chinese herbs. Comput Appl Chem 24:41-44
- Xu X (2006) New concepts and approaches for drug discovery based on traditional Chinese medicine. Drug Discov Today Technol 3:247-253
[Crossref] [Google Scholar] [PubMed]
- Xu X (1999) Study on computer simulation for Chinese traditional compound medicine. Prog Chem 11:202-204

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences