Advances in Triterpenoid Saponins Research 2007-2012
Saraswati Garai
DOI10.21767/2472-0151.100020
Saraswati Garai*
CSIR-Indian Institute of Chemical Biology, Jadavpur, Kolkata, West Bengal, India
- *Corresponding Author:
- Saraswati Garai
CSIR-Indian Institute of Chemical Biology
4 Raja SC Mullick Road, Jadavpur
Kolkata-700 032, India.
Tel: +913324995947
Fax: +913324735197
E-mail:saraswatigarai@iicb.res.in
Received date: November 14, 2016; Accepted date: November 29, 2016; Published date: December 05, 2016
Citation: Garai S. Advances in Triterpenoid Saponins Research 2007-2012. Herb Med.2016, 2:3. doi:10.21767/2472-0151.100020
Abstract
Triterpenoid saponins isolated and identified from natural resources during the period April, 2007-December, 2012 are reviewed. Current techniques used in their isolation and structure determination are discussed. New triterpenoid saponins isolated along with their occurrence, selected physical data, spectroscopic methods used for their identification and the 13C NMR data of the novel aglycones of the parent saponins are compiled. The biological properties of those molecules are also included. Green corrosion inhibition effects of saponins and biosynthesis of triterpenoid saponins and production of triterpenoid saponins by tissue culture of those molecules are also discussed.
Keywords
Triterpenoid saponins; Isolation; Structure elucidation; Biological activity; Biosynthesis; Corrosion inhibition
Introduction
Triterpenoid saponins occur widely throughout the plant kingdom, marine microorganisms and phytoplankton, algae, sponges, mollusks, mangroves and microorganisms. The chemical and structural diversity of saponins with useful bioactivities, challenges of identification, unusual biosynthetic process and chemical synthesis for agricultural, industrial, and pharmaceutical applications have generated greater interest for further exploration of saponin chemistry and biology. The triterpenoid saponins reported up to March, 2007 have been reviewed recently [1] This review presents a list of new triterpenoid saponins published during April, 2007-December, 2012 and arranged skeletonwise, discusses newer trends in identification of triterpenoid glycosides, and lists the 13C NMR data of the novel aglycones of the triterpenoid saponins together with the biological activities and green corrosion inhibition effects of saponins and biosynthesis of triterpenoid saponins and production of triterpenoid saponins by tissue culture of those molecules reported.
Isolation
Triterpenoid saponins occur as complex mixtures. The different extraction techniques currently used for separation and isolation are supercritical fluid extraction (SFE), pressurized liquid extraction (PLE), microwave-assisted extraction (MAE), solid-phase microextraction (SPME), ultrasound-assisted extraction (UAE), superheated liquid extraction, and extraction with supercritical or subcritical water. The recent advances in separation methods are semi preparative HPLC, medium pressure liquid chromatography (MPLC), high speed countercurrent chromatography (HSCCC) and flash chromatography [2]. Some examples are provided.
Erylosides from the caribbean sponge Erylus formosus were isolated as follows. The lyophilized specimens were macerated and extracted with EtOH. The dried EtOH extract was chromatographed at low-pressure using a hydrophobic column eluting with H2O and 50% EtOH. The polar glycosidic fractions were purified by flash chromatography on silica gel to obtain the saponin fractions. The most polar saponin fractions thus obtained were further purified by HPLC on a Si gel column followed by a reverse phase column [3]. Yuan et al. isolated three new antifungal triterpene glycosides from the sea cucumber Holothuria axiloga. The dried sea cucumbers were refluxed with 60% EtOH. The dried extract was partitioned thrice with water and n-BuOH. The n-BuOH extract was suspended in CHCl3-MeOH and filtered. The filtrate was chromatographed over silica gel. The saponin-containing fractions were combined according to their TLC behaviour. The combined fractions were further purified by reversed phase silica MPLC followed by HPLC using Zorbax 300 SB-C18 [4]. For Holothuria scabra, the powdered sea cucumbers were refluxed four times with 60% EtOH. The dried EtOH extract was applied to a DA101 column and eluted successively with H2O, 70% EtOH and 95% EtOH. The saponin fractions thus obtained were repeatedly separated by MPLC with a column of reversedphase silica followed by HPLC [5].
The cytotoxic triterpene glycosides from the Malagasy plant Physena sessiliflora were isolated by Inoue et al. [6]. The dried leaves were boiled with MeOH. After filtration, the extract was evaporated to dryness. The extract was partitioned between equal volumes of n-hexane and MeOH-H2O (7:3). Methanol was removed from MeOH-H2O layer. The aqueous layer was further partitioned with EtOAc and then n-BuOH successively. The n-BuOH fraction was chromatographed on silica gel and finally purified by HPLC and MPLC on an ODS column to give eight triterpene glycosides. The dried and powdered whole plants of Echinopsis macrogona were extracted with CHCl3 and MeOH under reflux. The MeOH extract was applied to a Diaion HP-20 column and washed with H2O, MeOH-H2O (3:7) and 100% MeOH. The MeOH eluted fraction was subjected to column chromatography with various solvent systems to give eight fractions. The fractions were further purified by repeated ODS and silica gel column chromatography using various solvent systems followed by HPLC to afford pachanosides C1, E1, F1 and G1, and bridgesides A1, C1, C2, D1, D2, E1 and E2 [7]. Gao et al. isolated four new triterpenoid saponins from Patrinia scabiosaefolia in another example. The dried plants were extracted with methanol overnight at room temperature. The MeOH extract was partitioned against EtOAc and H2O. The aqueous part was applied to HPD 450 macroporous resin column and washed with H2O and EtOH. The EtOH eluate was purified by vacuum low-pressure column chromatography on silica gel using CHCl3-MeOH in various ratios to afford seven fractions. Rechromatography of the fractions over ODS-C18 followed by repeated MPLC and further HPLC purification furnished four triterpenoid saponins [8].
Structure Elucidation
The introduction of hyphenated techniques like HPLC-UV, HPLCCAD, HPTLC, High-Speed counter current chromatography coupled with evaporative light scattering detection, or LCMALDI- TOF-MS allow simultaneous structure determination of novel biologically active saponins with a small quantity of material avoiding the time consuming isolation of pure components. Ma et al. isolated four oleanane-type triterpene saponins, esculentosides A, B, C and D from the roots of Radix phytolaccae by high speed countercurrent chromatography coupled with evaporative light scattering detection [9]. Several triterpenoid saponins from Pulsatilla koreana have been analyzed simultaneously by High Performance Liquid Chromatography coupled with a Charged Aerosol Detector (HPLC-CAD) [10]. The saponins of the medicinally important plant Aerva lanata were characterised by utilizing developed HPTLC finger prints. These fingerprints help the manufacturer to distinguish the adulterant and permit standardization of herbal formulations [11]. Triterpenoid saponins from the medicinal plant Maesa lanceolata were analyzed by LC-UV/MS system [12]. A rapid and sensitive bioassay based on liquid chromatography tandem mass spectrometry (LC-MS/MS) for the simultaneous determination of four isomeric escin saponins in human plasma has been developed and validated [13]. In order to obtain the clinically relevant saponin-related pharmacodynamics of the compound Danshan tablets composed of Panax notoginseng, a specific and sensitive LC-MS/MS method was developed for simultaneous determination of three bioactive saponins notoginsenoside R1 and ginsenosides Rg1 and Rb1 after a single oral administration [14]. The chemical transformation of 20(S)-protopanaxadiol-type ginsenosides Re, Rge and Rf in acidic conditions was analyzed by a rapid resolution liquid chromatography coupled with Quadrupoletime- of-flight mass spectrometry (RRLC-Q-TOF-MS) method [15]. A new liquid chromatography methodology coupled to a diode array detector and a time-of-flight mass spectrometer has been developed for the simultaneous determination of saponins in Chenopodium quinoa [16].
Mass spectrometry
The utility of HPLC-ESIMS in the structure elucidation of oleananetype triterpene glycosides was discussed by Gulcemal et al. [17]. In the MS/MS of [M-H]- ion of the oleanane bisdesmoside studied, product ions appeared at m/z 941 [M-H-Glc]-, 923 [M-HGlc- H2O]-, and 879 [M-H-Glc-H2O-CO2]- from the cleavage of glycosidic bonds and due to the presence of carboxyl gr. in the hexose sugar. The ESI-Q-TOF MS of besylvoside 1, an oleananetype triterpene monodesmoside, generated [M-H]- peak at m/z 1105.523, MS/MS of which yielded ions of m/z 487.283 and 617.139 indicating the presence of deprotonated aglycone and the glycone moiety (Scognamiglio et al.). The sequence of monosaccharide units in the carbohydrate chain of liouvillosides of the sea cucumbers Staurocucumis liouvillei was determined by (-) ESI-FTMS and CID MS/MS (Antonov et al.). The MS/MS of an [M2Na-2Na]2- of liouvilloside A1 afforded ions at m/z 987.42 [M2Na- MeGlc-NaSO3 +2H-Na]-, 781.38 [M2Na-MeGlc-Glc-NaSO3+H-Na- CO2]-, 563.13 [M2Na-Xyl-NaSO3-OAgI-H-Na]-, 387.06 [M2Na-OAgI- 2Na-H]-. It suggested that four of the sugars were present as a tetrasaccharide unit. The usefulness or application of 2D-HPLCESI- ITMSn in the structure determination of quillaja saponins was reported by Bankefors et al. In MS1/MS2 the mostly observed [M+Na]+ ion of the compound S1 provided fragments by loss of C-3 {[A+Na]+, i.e., [M+Na]+ - [GlcA+Gal+Na]+} and C-28 {[B+Na]+, i.e., [GlcA+Gal+Na]+ - [Fuc+Rha+Na]+} glycosidic bond cleavages. Furthermore, 43 new [B+Na]+ ions were found corresponding to new types of C-28 oligosaccharides in 143 components.
NMR spectrometry
The fast and multidimensional 2D, 3D and 4D NMR experiments and also Computer assisted structure elucidation programs have revolutionized for the structural studies of saponin without degradative chemistry.
The planar structure and the relative configuration of the aglycone are being determined by detailed analysis of COSY, HMBC and NOESY experiments. The HMBC correlation at δ (H) 6.22/ δ (c) 176.3 of yemuoside YM18 (1) indicated that the sugar chain was attached at C-28 through an ester bond. The correlations at δ (H) 4.95/ δ (c) 69.4 and δ (H) 5.83/ δ (c)78.3 confirmed the interglucosidic linkages. The coupling constants and chemical shifts of the anomeric H atom signals at δ 6.22 (J=8.1), 4.95 (J=7.6) and 5.83 (br, s) suggested the anomeric configurations of the sugars to be β -glucose, β -glucose, and α -rhamnose [18].

The structure of one of the four triterpenoid saponins with a novel aglycone isolated from Patrinia scabiosaefolia was established by spectroscopic methods and chemical transformations. The 13C NMR of Saponin (3) displayed 46 carbon signals including 30 for the aglycone part and the remaining 16 for the glycone part. The HSQC spectrum displayed a correlation between the C-13 signal and the carbonyl signal C-28 suggesting the presence of 28, 13 β lactone of the aglycone. The absence of one tertiary methyl group (C-30) which was replaced by CH2OH was confirmed by the upfield shift of C-29 and downfield shift of C-20 signals. The 1H NMR spectrum exhibited six tertiary methyl signals and another doublet methyl proton signal. HMBC correlations were observed between H-16/H-22 and C-28, H-15/H-18/H-27 and C-13, 2H-11 and C-12, H-19/H-21/3H-30 and C-29, and H-19/H-21/2H-29 and C-30, suggesting the lactone moiety connection of the aglycone. The NOESY spectrum correlation between H-12, H-9 and Me- 27 indicated the α -configuration of H-12. The 1H and 13C NMR disclosed the presence of one rhamnose and two xylose units. The configurations of the xylose (β) and rhamnose (α) were revealed from 3JH-1,H-2 which were found to be 8 Hz, 8 Hz and 6 Hz [19].

The structure of a new triterpenoid saponin, Gummososide (3) with a novel aglycone, isolated from a cacti, was determined as follows The 1H and 13C and DEPT NMR spectra identified the pachanane-type skeleton of the aglycone moiety attached with three sugars. It displayed seven methyl groups, eight methyl carbons, three anomeric signals, olefinic signals and two carbonyl signals. The linkage site of the three sugars, glucuronic acid, xylose and glucose were confirmed by HMBC and NOESY correlations. It disclosed that the glucuronic acid and glucose are 1, 2 linked and glucuronic acid and xylose are 1, 3 linked. The β -configuration of three sugars was deduced from NOESY. The HMBC correlation spectrum indicated an acetyl group at C-22 with α configuration as revealed by the NOESY spectrum [20].

Biological Activity
Saponin discovery research within the agriculture, industry and pharmaceutical industry have some technical limitations or drawbacks still now in the third world countries.
Antifungal activity
The triterpenoid saponins exhibit antifungal activities and prevent important crop loss in modern agriculture and horticulture. They exert strong antifungal activity against a broad range of fungi types and stages. Saponins prevent the synthesis of ecdysteroid, the protease inhibitors, or are cytotoxic to certain fungi. They inhibit fungal growth and replication. Saponins could be used exogenic by spraying them on fields. Saponin powder and solutions are already commercially utilized as natural insecticides in the third world countries with Integrated Pest Management (IPM) programs [21]. The structure-activity relationships of saponins need however to be further studied. Fungal infection to human health have been increasing worldwide. Natural saponins with novel skeletons have been stimulating great interest from drug discovery community for newer antifungal agents widely used in clinical practice [22]. The antifungal activity of holostanetype triterpenoid saponins of Sea cucumber Bohadschia marmorata was investigated against six strains: Candida albicans, Crystococcus neoformans, Aspergillus fumigatus (Clinical strain), Trichophyton rubrum, Candida tropicalis (Clinical strain), and Candida krusei (Clinical strain) to find 0.70 ≤ MIC80 ≤ 2.81μM [23]. The triterpenoid saponins isolated from Lecaniodiscus cupanioides showed antifungal activity against Candida albicans, Candida neoformans and Aspergillus fumigates [24]. The monodesmoside triterpenoid glycoside phytolaccoside B of Phytolacca tetramera showed antifungal activity in agar dilution assays in the absence and presence of different concentrations (50-250 μg/mL) of ergosterol to the assay medium. Triterpenoid saponins generally bind with membrane ergosterol causing fungal membrane disruption and loss of the intracellular content. The MIC of phytolaccoside B for S. cerevisiae was tested. The MIC remained unchanged, suggesting no binding to membrane ergosterol. The leakage effect has been tested in assay medium containing phytolaccoside B (74 μM), cultured cell suspension of S. cerevisiae and SDS (2%) to note inhibition of human opportunistic and pathogenic fungi. Phytolaccoside B damaged the fungal membrane by interacting with elements due to amphipathic nature. This study was also performed with human and mammalian erythrocytes. TEM and SEM studies showed morphological changes in yeasts and produced by enfumafungin in vitro (Table 1). The saponin did not inhibit (1 → 3)- β -D-glucan synthase and exhibited enhanced activity to arrest cell growth [25] (Table 2).
| Fungal spp | Voucher specimen | Phytolaccoside B | Amph | Ket | |||
|---|---|---|---|---|---|---|---|
| MIC100 | MIC80 | MIC50 | MFC | MIC100 | MIC100 | ||
| Candida albicans | ATCC 10231 | 74 | 74 | 74 | 150 | 0.84 | 1 |
| Saccharomyces cerevisiae | ATCC 9763 | 74 | 37 | 18 | 150 | 1.62 | 1 |
| Cryptococcus neoformans | ATCC 32264 | 112 | 74 | 9 | 112 | 0.27 | 0.5 |
| Neurospora crassa | ATCC 9279 | 74 | nt | nt | nt | 1 | 2 |
| Aspergillus niger | ATCC 9029 | 188 | nt | nt | nt | 0.5 | 0.25 |
| Aspergillus fumigates | ATCC26934 | 188 | nt | nt | nt | 0.5 | 0.13 |
| Aspergillus flavus | ATCC 9170 | 74 | nt | nt | v | 0.5 | 0.5 |
MIC100, MIC80, and MIC50: Concentration of a compound that caused 100%, 80%, or 50% reduction of the growth control, respectively. ATCC=American Type Culture collection(Peoria, IL): Amph=amphotericin B; Ket=ketoconazole; n.t=not tested.
Table 1: Minimum Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC) in μM of Phytolaccoside B.
| Compound | (1→3)-β-D-Glucan Synthase | Chitin Synthase 1Basal(-trypsin) |
|---|---|---|
| IC50 | IC50 | |
| Phytolaccoside B | 0.5 | >0.40 |
Table 2: In VitroAntigungal activity of Phytolaccoside B into insoluble (1→3)-β-D-Glucan and Chitin.
Immunomodulatory activity
Saponins have recently drawn much attention for their use in adjuvants for human vaccine. A review on saponin-based adjuvants is available [26]. Ginseng saponins from Panax ginseng have been identified as potent adjuvants. The adjuvant effects of the ginsenosides have been investigated against OVA in mice. The ginsenosides Rg1, Re, Rg2, Rg3 and Rb1 were found to enhance significantly the serum OVA specific IgG, IgG1 and IgG2a antibody titers in the immunized mice. Rg1 enhanced significantly splenocyte proliferative responses to ConA, LPS and OVA. Rg1 also increased maximum production of the cytokines IL-5 and IFN-γ (P<0.05) in OVA immunized mice. The molecular mechanism of adjuvants is the formation of the immustimulating complex (ISCOM), OVA-specific IgG2a/IgG1 productions via Th1/ Th2 cytokines activation and IFN- γ/IL-5 secretions [27]. The immune responses Th1/Th2 of the least hemolytic ginsenoside Rd from Panax notoginseng have been studied in mice against ovalbumin (OVA). It could significantly increase the activation potential of T and B cells in mice immunized with OVA. Rd significantly enhanced splenocyte proliferation and OVA-specific serum antibody response in immunized mice. The production of Th1 and Th2 cytokines was induced by interleukin 2 (IL-2), interferon-γ (IFN-γ), II-4 and IL-10 mRNA expression in mice splenocytes induced by ConA. The immunological activity was performed mechanistically by regulating production and gene expression of Th1 and Th2 cytokines [28].
The oral administration of ginseng stem and leaf saponins (GSLS) on intranasal immunization with a live Newcastle disease vaccine has been examined in chickens. Hemagglutination inhibition (HI) titer is increased maximally at a dose 5 mg/kg of GSLS for 7 days. Splenic peripheral lymphocyte stimulation response (p<0.01) was found along with more IgA+ cells in duodenum (p<0.05) and ilELS in jejunum (p<0.01). The ability of GSLS to augment both antibody and cell-mediated immune responses suggested that this adjuvant could be a useful oral adjuvant to improve vaccine immunization in chickens. This effect may be due to the antagonistic effect between ginsenosides [29].
GSLS alone or in combination with oil significantly enhanced immune response to Face and Mouth Disease vaccination (FMDV) in immunized mice. FMDV antigen with oil/GSLS induced significantly higher IgG and the subclass responses, and production of IFN-γ and IL-5 by splenocytes, and caused splenocyte proliferation in response to ConA and LPS than when used alone or mixed with either GSLS or oil. This result suggests that GSLS and oil synergistically promote both Th1 and Th2 immune responses. This synergism between GSLS and oil may result from the depot effect of oil emulsion in combination with the immunomodulatory effect of GSLS. For better immunity against FMDV infection in cattle and pigs, the combined effects of GSLS and oil in cellular and humoral immune responses may be studied further [30]. The usage of GSLS in inactivated FMD Alhydragel vaccine as an adjuvant has been evaluated through the estimation of cellular and humoral immune levels using lymphocyte blastogenesis assay, SNT and ELISA. It gave long lasting immunity and improved both cellular and humoral immunity. GSLS enhanced antibody production and acted as an activator of the Th1 response which was characterized by the production of antigen-specific IgCa a Th1 and the secretion of gamma interferon and interleukins which favour cellular immunity. It also enhanced interleukins which enhance cell mediated immune response [31].
The adjuvant roles of herbal medicines like ginseng and Salviae extracts on early immune responses during influenza virus infection have been investigated in a mouse model. The herbal extracts increased the levels of influenza virus specific antibodies and neutralizing activities when co-administered with inactivated virus. Salviae extract enhanced significantly IFN-γ and IL-2 cytokine responses after challenge infection. Ginseng induced high levels of IL-4 and IL-5 cytokine producing lymphocytes. The herbal extracts modulated leukocytes expressing CD69, an early activation marker and induced high levels of lung IgA antibody after challenge infection. The mechanism of adjuvant and potential immunomodulator of the herbal extracts is unknown [32]. The immunomodulatory properties of saponins of Tribulus terrestris on in vitro phagocytosis by murine peritoneal macrophages have been evaluated with regard to non-specific cellular immune responses. The saponin fractions showed highest phagocytic activity at dose 200 μg/ml. This indicated that the saponins enhanced the phagocytic efficacy of the PMN cells by causing more engulfment of the yeast cells, thereby stimulating a non-specific immune response. As neutrophils form the first line of host defence by virtue of their ability to phagocytose invading microorganism and foreign body, they have a major role in modulating the immune function. The stimulation of neutrophils results in an increase in the immediate non-specific cellular immune response. The augmentation of the host immune system may appear to be mediated through direct activation of macrophages and a possible stimulation of nonspecific immunity at low and non toxic doses [33].
Cytotoxicity of saponins
Saponins have been found to possess significant anticancer properties. Recently clinical studies of saponins have been highlighted [34-36]. Natural saponins are attractive chemopreventive agents due to their very low clinical toxicity compared with chemical antitumor drugs. Tested against the murine macrophage-like cell line RAW264.7, a 5:4 mixture of two bisdesmosidic saponins of the aglycone 16 α -hydroxyprotobassic acid, obtained from the roots of S. foetidissimum, showed stronger cytotoxicity with IC50 values at 11.9 ± 1.5 μg/ml than the saponin fraction (IC50=33.7 ± 6.2 μg/ml) of the ethanol extract. The cytotoxic activity is suggested to be due to the bisdesmosidic nature of the saponins [37]. The saponin fraction from Pulsatilla chinensis inhibited cellular proliferation of 7402 cells induced by apoptosis and accumulation of cells at S-phase and Sub-Go phase (Cell death) in a dose dependent manner. It also inhibited the growth of human liver cancers in nude mice in xenograft models. The number of leukocytes, and leukomonocyte and neutrophil percentages in peripheral blood showed almost no significant change between the PRS and control groups, while CTX are markedly reduced in the number of leukocytes. These results indicated that PRS had no marked toxic side effect as revealed by the number of leukocytes and leukomonocytes in peripheral blood. The weight loss needs to be further investigated due to no significant pathologic changes in organ tissue. The mechanism of antitumor action is unknown. So, the saponins of P. chinensis may be utilized as chemotherapeutic agents in human lung cancer [38].
A single saponin from Gypsophila paniculata exhibited strong cytotoxicity enhancing properties on Sap-EGF on HER14 cells in a wide range without causing self-cytotoxicity. It increased the cytotoxicity of Sap-EGF drastically with an enhancement of about 4900 fold (IC50: 0.0008 nM) which was lower than the SA-induced enhancement. The IC50 of Sap-EGF alone was 2.47 nM [39]. The three triterpene saponins saikosaponin a, buddleja saponin IV and songarosaponin D from Physospermum verticillatum exhibited remarkable cytotoxic activity against lung large carcinoma COR- 123 cell line, with 90 times stronger effect than the positive control. Buddlejasaponin IV and songarosaponin D exerted significant inhibition of NO production in LPS induced RAW 264.7 macrophages with IC50 of 4.2 and 10.4 μM. The saponins did not affect the proliferation of skin fibroblasts 142BR suggesting a selective action against cancer cells. The mechanism of the chemopreventive activity of saponins is the ability to reduce oxidative stress responsible for the release of different mediators such as NO. The interaction between levels of NO and several aspects of tumor development needs to be further studied. The saponins from P. verticillatum may be good candidates for potential antitumor compounds interfering with NO production. They may also have potential value for the design of novel semisynthetic analogues with chemopreventive and/or cytotoxic activity [40]. The production of cycloartane saponins from hairy roots of Astragalus membranaceus in airlift bioreactor has been extended for human clinical studies to evaluate their cytotoxic and apoptosis induction potentials in a panel of human tumor cell lines [41]. The metabolite compound K of Protopanaxatriol of Panax ginseng was found to inhibit the viability of HL-60 human leukemia cells in a dose and time dependent manner with an IC50 of 14 μM. This cell death had typical features of apoptosis that is DNA fragmentation, DNA ladder formation and the externalization of annexin V targeted phosphatidylserine residues in HL-60 cells. Compound K induced a series of intracellular events associated with both the mitochondrial and death receptor-dependent apoptotic pathways, the activation of caspases-3, -8, and -9, the loss of mitochondrial membrane potential, the release of cytochrome c and Smac/DIABLO to the cytosol, and the translocation of Bid and Bax to mitochondria. The downregulation of Bcl-2 and Bcl-Xl, Caspase-8 inhibitors, by Compound K completely abolished caspase-3 activation, Bid cleavage, and subsequent DNA fragmentation. The activation of caspase-3 and -8 and DNA fragmentation were significantly prevented in the presence of cycloheximide, suggesting that compound K induced apoptosis is dependent on de novo protein synthesis. These results indicated that caspase-8 plays a key role in compound K stimulated apoptosis via the activation of caspase-3 directly or indirectly through Bid cleavage, cytochrome c release, and caspase-9 activation [42]. The cytotoxic mechanism of compound K with respect to the involvement of reactive oxygen species (ROS) and the mitochondrial involved apoptosis in HT-29 human colon cancer cells has been studied. It demonstrated that Compound K-mediated generation of ROS led to apoptosis through activation of p38MAPK and JNK which modulate the expression of Bcl-2 and Bax, and then trigger loss of mitochondrial membrane potential, cytochrome c release and caspase activation [43].
Prostate cancer is a major public health problem worldwide. The 20(S)-ginsenoside Rh2 inhibited proliferation of both androgendependent and independent prostate cancer cells. The 20(S)- hydroxy group contributed to the cytotoxic activity [44]. D-Rh2, the dioctanoyl ester of Rh2 could significantly decrease toxicity to the human hepatocyte cell line QSG-7701 in vitro but not attenuate the antitumor activity in vivo compared with Rh2 (Figure 1) and could be a more potential candidate for use as an antitumor drug due to the possible lower side effect [45].
The differential cellular uptakes of ginsenosides from white ginseng and red ginseng and their effects on the proliferation and apoptosis in human breast cancer cells have been studied. Protopanaxadiol from red genseng exhibited higher cellular uptake in MCF-7 cells than protopanaxatriol from white ginseng which explains the difference in cell proliferation. PPD, but not Rg1, was found to induce substantial apoptosis through the activation of caspase-3. This suggested that the amount cellular uptake of ginsenosides is a critical component for their cellular actions. RG exhibited the most potent cytotoxicity at 50 to 100 μg/ml dose range on MCF-7 and MDA-MB231 cells. RG exerted more potent cytotoxic and apoptotic effects than WG against MCF-7 and MDA-MB231 cells. PPD from RG was found to be the more potent agent and needs to be explored further as a potential anticancer agent for breast cancer treatment [48].
Ginsenoside Rd inhibited the expression and activation of MMP- 1, MMP-2, and MMP-7 by inactivating ERK1/2, p38MAPK, and JNK signaling pathways and inactivation of AP-1 activation. It inhibited the invasion and migration of HepG2 cells in vitro by activating of E-cadherin and vinculin and decreasing the level of N-cadherin. Ginsenoside Rd may be a new candidate for an anticancer agent [46].
The effect of main intestinal bacterial metabolite of ginseng (MI, PPD) on inflammation-related metastasis of colon 26 cells has been investigated using an in vitro TNF- α stimulation model. MI suppressed TNF- α enhanced metastasis by downregulating NFxB signaling in colon 26 cells. In TNF- α treated colon 26 cells, MI suppressed the TNF- α induced expression of MMP-9 at the level of both mRNA and protein, and the production of TNF- α in peritoneal macrophage and spleen cells. The cytokine stimulation model might facilitate the evaluation of saponins effective in vivo experimental models [47].
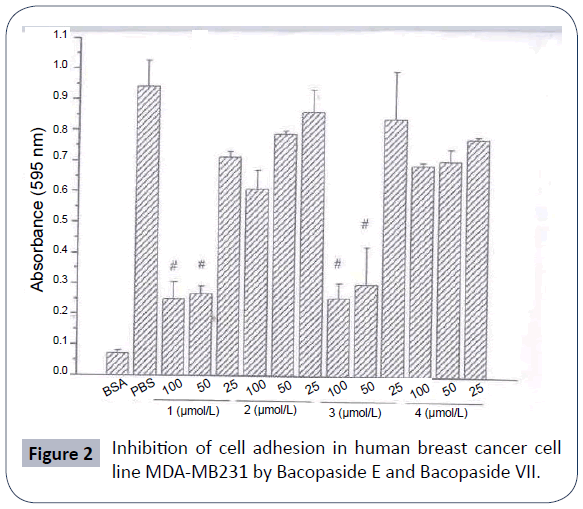
The methanol extract of Bacopa monniera was found to exhibit cytotoxicity against human tumor cell lines. The two dammarane triterpene saponins, bacopaside E and bacopaside VII of B. monniera n-butanol fraction exhibited cytotoxicity at the concentration of 50 μmol/kg against human tumor cell lines MDA-MB-231, SHG-44, HCT-8, A-549 and PC-3M in MTT assay in vitro and showed 90.52% and 84.13% inhibition in mouse implanted with sarcoma S180 in vivo (Table 3). They also inhibited significantly the adhesion, migration and matrigel invasion of human breast cancer cell line MDA-MB-231 in vitro (Figures 2-4). The mechanism of cytotoxicity needs further study. The purified bacopasaponin fractions of B. monniera may be potential antitumor agents due to their dammarane skeleton [48]. Bacoside A from Bacopa monniera has a potent antimetastatic effect against DEN-induced hepatocellular carcinoma in rats by reducing the activities and expression of MMP-2 and MMP-9. It could be an efficient therapeutic agent in the inhibition of cancer development/metastasis [49].
| Saponin | IC50(μmol/L) | Dose | Inhibition ratio | ||||
|---|---|---|---|---|---|---|---|
| MDA-MB-231 | SHG-44 | HCT-8 | A-549 | PC-3M | (μmol/kg) | (%) | |
| Bacopaside E | 12.3 | 9.7 | 11.3 | 8.9 | 13.9 | 50 | 90.52 |
| Bacopaside II | 32.4 | 36.9 | 40.3 | 44.4 | 45.4 | 50 | 16.27 |
| Bacopaside VII | 14.3 | 15.9 | 9.8 | 9.7 | 10.1 | 50 | 84.13 |
| Bacopasaponin C | 34.9 | 34.9 | 47.3 | 56.4 | 48.2 | 50 | 35.51 |
Table 3: In vitrocytotoxic against human tumor cell lines and inhibition ratio of sarcomaS180 in mouse.
Antimicrobial activity
Antimicrobial activity of the methanolic extract of leaf callus of Bacopa monniera has been investigated against four bacterial strains Bacillus subtilis, Staphylococcus aureus, Pseudomonas aeruginosa, and Klebsiella pneumoniae, and four fungal strains Microsporum audovini, Trichophyton mentagrophytes, Candida albicans, and Aspergillus niger at concentrations 0.03, 0.05, 0.12 and 0.25 mg/disc (Tables 4 and 5). It showed significant and dose dependent inhibition activity compared to the standard drug ciprofloxacin/Griseofulvin [50].
| Leaf callus ext. Concentrations (mg/disc) | Diameter of zone of inhibition (mm) | |||
|---|---|---|---|---|
| Fungal strains | ||||
| Candida albicans | Microsporum audouinii | Aspergilus niger | Trichophyton mentagrophytes | |
| 0.03 | 4.4 | 5.2 | 3.8 | 5 |
| 0.05 | 5.8 | 7.1 | 5.5 | 7.3 |
| 0.12 | 10.2 | 11.5 | 9.2 | 10.5 |
| 0.25 | 12.5 | 13.8 | 11.2 | 13 |
| Control (DMSO) | ||||
| Griseofulvin (0.25) | 11.3 | 12 | 10.5 | |
Table 4: Antibacterial activity of extract of leaf callus of Bacopa monniera L.
| Leaf callus ext. Concentrations (mg/disc) | Diameter of zone of inhibition (mm) | |||
|---|---|---|---|---|
| Fungal strains | ||||
| Candida albicans | Microsporum audouinii | Aspergilus niger | Trichophyton mentagrophytes | |
| 0.03 | 4.4 | 5.2 | 3.8 | 5 |
| 0.05 | 5.8 | 7.1 | 5.5 | 7.3 |
| 0.12 | 10.2 | 11.5 | 9.2 | 10.5 |
| 0.25 | 12.5 | 13.8 | 11.2 | 13 |
| Control (DMSO) | ||||
| Griseofulvin (0.25) | 11.3 | 12 | 10.5 | 11.5 |
Table 5: Antibacterial activity of extract of leaf callus of Bacopa monnieraL.
The antibacterial activity of Chlorophytum borivilianum (roots and stem) glacial acetic acid extract against Staphylococcus aureus, Bacillus subtilis, Escherichia coli and Pseudomonus aeruginosa has been studied. It exhibited very good antibacterial effect. The main constituents mannans and saponins form a mucilaginous layer around the urinogenital, gastrointestinal and respiratory tract after oral administration. The bacteria cannot attack the system due to the formation of layers which trap microbial flora [51]. The acidic methanol extracts of Gymnema sylvestre have been evaluated for antimicrobial activity against S. aureus and S. typhimurium. It showed good activity towards the pathogens. The broad spectrum activity of the plant can be utilized in the development of new antimicrobial drugs [52]. The saponin fractions of Gymnema sylvestre and Eclipta prostrata have been evaluated for antimicrobial activity against Bacillus pumilus, Bacillus subtilis, Pseudomonas aeruginosa and Staphylococcus aureus in agar diffusion assay (Table 6).
| Microorganisms | Crude saponin zone of inhibition (mm) (10,000 mg/l) | MIC(mg/l) | Pure saponin zone of inhibition (mm) (10,000 mg/l) | MIC(mg/l) | ||||
|---|---|---|---|---|---|---|---|---|
| G.S | E.P | G.S | E.P | G.S | E.P | G.S | E.P | |
| Pseudomonas aeruginosa | 12 | 8 | 1200 | 1200 | 20 | 20 | 800 | 1200 |
| Escherichia coli | 10 | 16 | 1200 | 1200 | 16 | 16 | 1000 | 1200 |
| Salmonella typhi | 14 | no activity | 1200 | no activity | 16 | 16 | 1000 | 1200 |
| Klebsiella pneumonia | 14 | 8 | 1200 | no activity | 10 | 14 | 1200 | 1000 |
| Proteus mirablis | 14 | 16 | 1200 | no activity | 14 | 14 | 600 | 1000 |
| Staphylococcus aureus | 10 | 10 | 1000 | no activity | 19 | 8 | 1200 | |
| (20,000mg/l) | MIC(mg/l) | (20,000mg/l) | MIC(mg/l) | |||||
| Aspergillus fumigates | 20 | 16 | 2000 | 20 | 16 | 1400 | 1400 | |
| Aspergillus flavus | 8 | 16 | 2000 | 16 | 18 | 1400 | 1400 | |
| Aspergillus niger | 12 | 14 | 1600 | 16 | 22 | 1400 | 1400 | |
Table 6: Antimicrobial activity of crude and pure saponin fractions from the leaves of G.sylvestreand E.prostrata.
The MIC values of pure saponin fraction of G. sylvestre ranged from 600 to 1200 mg/l for tested bacterial pathogens [53]. The saponin rich Guar meal fractions exhibited antibacterial activity in a dose dependent manner when tested against two gram positive bacteria (Lactobacillus spp. and Staphylococcus aureus) and two gram negative bacteria (Escherichia coli and Salmonella typhimurium). Interest in the use of metabolic engineering and post genomics to plants for antimicrobial drug development has been increasing. But the characterization of mechanism of action of antimicrobials is still unknown [54].
The antimicrobial effects of scoposides A-E have been reported using the MIC method. Scoposide E was found to be most active for both Gram positive and Gram negative bacteriam and showed equal activity with gentamicin against E. faecalis. Scoposides A-D showed moderate activity [55].
Miscellaneous effects
Obesity and overweight are medical conditions in which excess body fat has accumulated to the extent that it may have an adverse effect on health which leads to decrease longevity and increased health problems like cardiovascular diseases, diabetes, cancer and respiratory problems. It is a global problem. Ginsenosides of Panax notoginseng may prevent high fat dietinduced fat storage in adipose tissue by inhibiting the intestinal absorption of dietary fat through the inhibition of pancreatic lipase activity. Ginsenoside Rc is potent pancreatic lipase inhibitor [56]. Protopanaxadiol type ginsenosides inhibited the pancreatic activity in a dose dependent manner at the concentrations of 0.25-1 mg/ml. They also inhibited hydrolysis of about 83.2% of triolein at about 1 mg/ml of PDG. PDG is effective in preventing and healing obesity, fatty liver and hypertriglyceridemia in mice fed with a high-fat diet [57]. The active saponins of Camellia sinensis suppressed the appetite signals in the hypothalamus through stimulation of the capsaicin-sensitive sensory nerves, probably vagal afferent nerves, or enhancement of 5-HT release from the ilea, leading to reduced food intake and body weight gain [58].
Leishmaniasis is a major global problem. The triterpenoid saponin, dasyscyphinic C of Eclipta prostrata was found to be more effective than gymnemagenol of Gymnema sylvestre in inhibiting the growth of L. major promastigotes [59]. The saponin adjuvant elicited strong antigenicity related to increased immunoglobulin isotypes, together with higher levels of lymphocytes, particularly of circulating CD8+ T-lymphocytes. The intense cell proliferation and NO production during in vitro stimulation by SLcA shows that LBSap vaccine elicited a potential immune activation status potentially compatible with effective control of the etiological agent of CVL. The efficacy of the LBAap vaccination in protection against experimental challenge with Leishmania chagasi demands further research [60].
Osteoclastic bone resorption has been increasing in the pathogenesis of many bone diseases. Osteoclast inhibitors are the most widely used treatments for these diseases. 20(R)-Rh2 ginsenoside showed a stronger inhibitory effect on osteoclast formation than 20(S)-Rh2 ginsenoside. But the latter is a stronger cytotoxic agent than the former on pre-osteoclast proliferation. The 20-hydroxyl group contributed to osteoclast formation and cytotoxicity [61]. Ginsenoside Rd induced differentiation and mineralization of osteoblastic MC3Te-E1 cells via the activation of AMPK and the subsequent enhancement of BMP-2 production. It might be beneficial for the treatment of bone diseases such as osteoporosis and for bone repair by promoting bone formation [62].
Bacopa monniera extract exhibited a protective effect against morphine-induced liver and kidney toxicity (Sumathi et al.). The triterpenoid saponins platycodins A, C and D and deapioplatycodin D isolated from Platycodi radix showed inhibitory effects on glutamine-induced toxicity in primary cultured rat cortical cells [63]. A protopanaxadiol-type ginsenoside and the metabolite compound K (Ginseng) decreased fasting blood glucose and plasma TG levels in type 2 diabetes induced by HFD/STZ via down-regulation of PEPCK and G6Pase expression in liver [64]. Polygalasaponin γ, bayogenin-3-O- β -D-xylopyranosyl (1 → 4)- α -L-rhamnopyranosyl (1 → 2)- β -D-glucopyranosyl ester of Polygala japonica, strongly inhibited acute inflammation in vivo. Bayogenin-3-O- β -D-glucopyranoside was found to significantly inhibit NO production in LPS-stimulated RAW264.7 macrophages. It explains the potential therapeutic utility of P. japonica saponins in traditional Chinese medicine (Wang et al.). The oleanane-type triterpene saponins from the bark of Aralia elata can be used to prevent inflammatory diseases. They affects metabolic function in human HepG2 cells. Elatoside L significantly reduced TNF α -induced iNOS and COX-2 gene expression in a dose dependent manner [65]. The hepatoprotective activity of bacoside A of Bacopa monniera against D-GalN induced liver injury in rats was examined. It reduced the elevated levels of serum alanine transaminase (ALT), aspirate transaminase (AST), ALP, γ-GT, LDH and 5′ND. It also significantly restored the decreased levels of Vit-C and Vit-E induced by D-GalN normal both in liver and plasma. Bacoside A of BM has hepatoprotective effect against D-GalN induced hepatotoxicity in rats (Sumathi et al.). Hepatitis B and C virus is a major global health problem. To control HCV and HBV, herbal drugs could be an alternative approach. Saponins have been reported to have anti-herpes simplex virus type1 (HSV-1) effect. HCV infectivity was significantly suppressed by 5 μg/ml of saponin. It indicated that the saponin inhibits HCV propagation. The saponin potentiates IFN- α -induced anti HCV activity in a dose dependent manner. The inhibition of HCV replication suggests that SOCS2 protein level was increased and SOCS3 gene expression was decreased by saponin in replication cells. SOCS2 is a negative regulator in HCV propagation. Thus the saponin may be a potent therapeutic agent for HCV patients [66].
The triterpenoid saponin 2 α, 3 β,19γ-trihydroxyurs-12-en- 28-oic acid β -D-glucopyranosyl ester from Potentilla anserine could decrease the expression levels of HBsAg, HBeAg and HBVDNA in the 2.2.15 cell culture. The saponin could be a potent therapeutic agent for HBV treatment [67]. Bacoside A relieved the oxidative stress induced by N-nitrosodiethylamine (DEN) in rats by decreasing LPO through scavenging of free radicals and enhancing the activity of antioxidants which could detoxify free radicals. The protective effect of bacoside A on N-nitrosodiethylamine-induced oxidative stress may be involved in free radical scavenging mechanism [68].
Green Corrosion Inhibition Effects of Saponins
Corrosion is a continuous process of oxidation of metal chemically or environmentally. Metal corrosion causes a great loss economically in various industries, e.g., oil, fertilizer, water treatment plants, pharmaceutical, chemical, aviation and aerospace. To prevent metal corrosion, numerous studies have been undertaken. Corrosion reactions are electrochemical in nature. They involve the transfer of charged ions across the surface between a metal and the electrolyte solution in which it is immersed. The organic and inorganic compounds as corrosion inhibitors are widely used to control metal corrosion as well as dissolution and acid consumption. These inhibitors are used in various industrial process such as acid pickling, acid cleaning, acid descaling and oil and oil well acidizing. Most of these inhibitors are highly toxic to both human being and living organism in environment, very expensive and nonbiodegradable. Due to increasing environment awareness, great attention has been paid by the global scientific community for green corrosion inhibitors, which are low toxic, biodegradable, low cost, readily available from renewable sources, ecofriendly and efficient. Plant products are environmentally friendly and ecologically acceptable. The recent work on plant extracts as corrosion inhibitors has major limitations due to lack of phytochemical investigations. Corrosion inhibition is a surface process. Various mechanisms of their action have been proposed for the corrosion inhibiting properties depending on metal, the media, and the structure of the inhibitor. Two modes of adsorption that occur as a result of electrostatic interaction between the green inhibitors and the metal surfaces are physical and chemical adsorption. The alcoholic extract of Bacopa monniera contains several dammarane-type, mainly jujubogenin and pseudojujubogenin triterpenoid saponins. Corrosion inhibition of bacopasaponins on mild steel in 1M HCl solution was studied. The maximum observed inhibition efficiency is 95%. The results obtained from both gravimetric and potentiodynamic polarization techniques (electrochemical techniques, XPS, Raman spectroscopy, FTIR, SEM) are very similar. Bacopasaponin C showed good corrosion inhibition efficiency and behaved as mixed type inhibitor. The inhibition efficiency of the bacopasaponin C increased with increase of concentration and reached highest value (95%) at 200 ppm. The evaluation of structure-activity relationship between the inhibitor and mild steel surface was performed. The adsorption of the inhibitor on mild steel surface is exothermic, spontaneous and consistent with the mechanism of physisorption. The empty p-orbital of oxygen atoms of triterpenoid saponins form bonds with the d orbital electrons on the metal surface (Garai et al.). The inhibitive and adsorptive characteristics of stem extract of Bacopa monniera in aqueous 1M NaOH solution for the corrosion of aluminium have been investigated. The extract showed maximum 75% corrosion inhibition efficiency at 400 mg/l [69]. Ginseng saponins are mainly protopanaxadiol and protopanaxatriol glycosides. The anticorrosive effects of ginseng extracts were studied for aluminium alloy of type AA 1060 in HCl solution. The crude extract exhibited the maximum inhibition efficiency with 93.1% at 50% v/v concentration. The inhibition efficiency increased with addition of iodide ions to the ginseng extract and the maximum observed inhibition was 96% at 30°C. The mechanism for the dissolution of Al metal and also hydrated cation and anion present on the metal surface has been proposed. Saponins have an ability to coordinate the metals via the d-orbitals of metal and empty p-orbitals of the oxygen atoms in the saponin molecules. Iodide ions have smaller degree of hydration, i.e., synergistic effect. They could bring excess negative charges in the vicinity of the interface, i.e., chemisorptions, and favour more adsorption of the positively charged saponin and negatively charged metal surface. This indicated that the enhanced inhibitory activity of ginseng extract observed might be due to the synergistic effect. The negative values of ΔGads0, ΔHads0 and ΔSads0 suggest that the adsorption of ginseng extracts onto aluminium surface is a spontaneous and exothermic process. The activation energy Ea, the enthalpy ΔHads0 and the entropy ΔSads0 of activation showed association of the saponin molecules of the ginseng root onto the aluminium surface rather than dissociation, i.e., physical adsorption [70]. The study of corrosion involves the study of the chemical, physical, metallurgical and mechanical properties of materials as it is a synergistic phenomenon in which the environment is as equally important as the materials involved. Computer modeling techniques can handle the study of complex systems such as corrosion and thus are appropriate and powerful tools to study the mechanism of action of corrosion and its inhibitors. The inhibitive and adsorption properties of alkaline crude extract of Phyllanthus amorus for both the corrosion and kinetics of corrosion process of aluminium in 2M sodium hydroxide solution were studied. The inhibition potential is attributed to the presence of saponins and other organic constituents. The maximum inhibition efficiency observed was 76% with 20% v/v of extract. This indicated that extract constituents are adsorbed onto the aluminium surface resulting in the blocking of the reaction sites and protects the aluminium surface from the attacks of the aggressive OH(-) ion from the alkaline solution [71]. The corrosion inhibition properties of crude extract of saponins of Newbouldea leavis leaf on aluminium alloy (AA8011) in 0.5M H2SO4 was studied. The crude saponin extract exhibited the maximum inhibition efficiency with 92% at the concentration of 400 mg l-1. The inhibition efficiency of aluminium in 0.05M H2SO4 increased as the extract concentration and temperature increased. The adsorption behavior of the inhibitors on the metal surface was investigated [72]. Ji et al. investigated the inhibitive effect of crude Argemone mexicana plant extract containing saponins on mild steel in 1M HCl. The observed maximum inhibition efficiency was 92.5% at a concentration 500 mgL-1 of the extract. The results indicated that the saponin extract in 1M HCl acted as a low cost and good anticorrosive agent. Surface morphology was studied by SEM. UV-visible, Scanning Electron Microscope (SEM), and Electrochemical impedence spectroscopy (EIS) were carried out for structure-activity relationship between the inhibitors and the mild steel surface. The visible UV studies suggests that the corrosion inhibition of the AM extract for the mild steel in HCl occurs through molecular adsorption on the metal surface. The results of EIS studies indicated that the crude extract of AM increased polarization resistance (Rp) and Rct and decreased Cdl, suggesting the adsorption of the crude saponins extract onto the mild steel surface. In SEM expts. the surface coverage parameter ÃÆââ¬ÅÃâè was increased with increasing inhibitor concentrations. The inhibitor molecules increased the surface coverage so that HCl could not react effectively with the mild steel and the metal corrosion was inhibited. Corrosion takes place through two reactions: cathodic (hydrogen evolution) reaction and anodic (metal dissolution) reaction. The big saponin molecules present in the extract inhibited corrosion probably by controlling both the anodic and cathodic reactions. Saponins are large molecules having triterpenes or steroids or alkaloid moiety and large sugar chains. These macro molecules have strong affinity toward metals and in acidic environment protonation takes place which probably shows better adsorption over the metal surface. These protonated species adsorbed on the cathodic sites of the mild steel and decreased the evolution of hydrogen. The adsorption on anodic sites probably occurred through lone pair electrons of oxygen atoms in the saponin molecules which decreased anodic dissolution of mild steel [73]. The corrosion inhibitive action of the aqueous extract of the leaves and stems of Anacyclus pyrethrum for mild steel in 0.5M H2SO4 has been investigated. The maximum inhibition efficiency was 87%. The inhibition also increased by the addition KI to LS-AP extract due to a synergistic effect. The decrease of the activation energy in the presence of LS-AP extract might be due to the chemisorptions of LS-AP extract inhibitor on mild steel involving charge sharing or charge transfer from the macromolecule saponins of LS-AP extract to the mild steel surface to form a strong coordinate-type bond. The increase in the polarization resistance (Rp) in the presence of LS-AP extract is due to the corrosion protection effect of the organic molecules. The decrease in the Cdlvalue in the presence of LS-AP extract indicated the adsorption of extract molecules on the metal surface. Surface morphology of the LS-AP extract were studied by Scanning electron microscope. A thin layer of the inhibitor molecules on the metal surface to prevent corrosion was observed [74]. Thus, saponin molecules may be used as low cost and best green anticorrosive agent in various industries.
Biosynthesis of Triterpenoid Saponins and Production by Tissue Culture
A functional genomic approach in the biosynthetic studies of saponins have been focused on the identification of enzymes involved. For understanding the two steps of monodesmoside formation, the biochemical genetics of saponin biosynthesis in Saponaria vaccaria has been studied through the identification and characterization of cDNAs encoding β -amyrin synthase (BAS) and an ester forming triterpene glucosyltransferase. The steps in the biosynthetic pathway towards saponins consist of the cyclization of 2, 3-oxidosqualene by BAS via the chair-chair-chair conformation, oxidation of β -amyrin at 16, 23 and 28 positions, glycosylation at positions 3 and 28 for the major bisdesmosides, and the acylation of sugars with acetyl and 2-hydroxy-2- methylglutaryl moieties. The enzymes involved in oxidation of β -amyrin may include cytochrome P450 and other hydroxylases, and alcohol and aldehyde dehydrogenases. Monodesmosides are formed by stepwise transfer of activated glucosyl donors at C-28 and catalyzed by the enzyme UDP-Glc-UGT74M1. The cloning of cDNAs encoding BAS and UGT74M1 provides some insights into saponin biosynthesis in S. vaccaria. The characterization of the UGT74M1 product indicated that it is a triterpene carboxylic acid glycosyltransferase. In vitro, the enzyme is capable of glycosylating a variety of oleanane triterpenes but has low activity with the lupene triterpenoid betulinic acid. NMR analysis of the glucosylation product of gypsogenin indicated that it forms a Glc ester at C-28. Vaccarosides A-D are monodesmosides having gypsogenic acid as the aglycone and a Glc linked to the carbonyl at C-28. The bisdesmosides, vaccarosides E-H, have a Fuc esterified to C-28 in addition to GlcUA at C-3. Based on experiments with GDP-Fuc and a variety of aglycones, UGT74M1 does not appear to be involved in making the Fuc ester linkage. Thus, UGT74M1 is specific for UDP-Glc and involved in the formation of monodesmosides. To elucidate the enzymes and genes involved in other steps in the biosynthesis of saponins in S. vaccaria further studies are required [75].
Most of the steps in the biosynthesis of triterpene saponins remain uncharacterized at the molecular level. The ongoing genome sequences in Medicago truncatula have been searched. The biosynthesis of triterpene saponins occurs through the initial cyclization of 2,3 oxidosqualene via the chair-chair-chair conformation by 2,3 oxidosqualene cyclases into potential dammarane, lupeol, α -amyrin or β -amyrin type. This cyclization is catalyzed by dammaranediol synthase, lupeol synthase (Lus), α -amyrin synthase, or β -AS respectively. Eight genes showing unique expression patterns and similarity to known 2,3-oxidosqualene cyclases are present in the Medicago genome sequence. The carboxyl group at C-28 position of the sapogenins in M. truncatula may be glycosylated by UGT73F3. NMR analysis inferred that the glucoside produced by the action of UGT73F3 with hederagenin as sugar acceptor was the C-28 ester [76]. In Medicago truncatula, a Cytochrome P450 gene (CYP716A12) has been identified in hemolytic saponin biosynthesis. A combined genetic and biochemical approach allowed to identify a cytochrome P450 gene (CYP716A12) for saponin biosynthesis. Genetic loss-of-function analysis and complementation studies showed that CYP716A12 is responsible for an early step in the saponin biosynthetic pathway. Mutants in CYP716A12 were unable to produce hemolytic saponins and only synthesized soyasaponins, and were thus named lacking hemolytic activity (lha). In vitro enzymatic activity assays indicated that CYP716A12 catalyzed the oxidation of β -amyrin and erythrodiol at the C-28 position to yield oleanolic acid. Transcriptome changes in the lacking hemolytic activity showed a modulation in the main steps of triterpenic saponin biosynthetic pathway, squalene cyclization, β -amyrin oxidation and glycosylation. The analysis of CYP716A12 expression in planta is reported together with the sapogenin content in different tissues and stages [77]. Avenacins, the antimicrobial triterpene saponins of oat roots, have been biosynthesized. The avenacin gene cluster have been also discussed. The β -amyrin synthase Sad1 catalyzed the formation of β -amyrin from 2,3-oxidosqualene in the biosynthetic pathway. Cytochrome P450 monooxygenase Sad2 is involved in the oxidation of β -amyrin. The Sad3 and Sad4 mutants have a significant role in avenacin synthesis and sequestration. The glucosyltransferase Sad3 and Sad4 gene products are involved in the addition of the β -1,4-linked D-glucose molecule to the triterpene moiety of avenacin A-1. The observed root morphological defects in both Sad3 and Sad4 mutants were due to mutations in genes which affect root growth and development. This implied the accumulation of monodeglucosyl avenacin A-1 in reduced root phenotypes [78]. The biosynthesis of novel and pharmacologically important triterpenoid saponins of Panax ginseng is an attractive strategy to achieve a higher medicinal value. The two squalene epoxidase genes PgSQE1 and PgSQE2 have been investigated for the transcriptional activity. Meja treatment enhanced the accumulation of PgSQE1mRNA in roots and prevented PgSE2mRNA. This suggests that the two genes are differentially regulated. The conversion of the precursor squalene to 2,3 (S) oxidosqualene was catalyzed by SQE genes. RT-PCR analysis indicated that squalene enhanced the accumulation of PgSQE1 and PgSQE2, SQS (squalene synthase), DDS (dammaranediol II synthease), PNY and PNX (cycloartenol synthase), suggesting the accumulation of ginsenosides in a dose dependent manner in in vitro cultured ginseng root. The RNA interference of PgSQE1 and PgSQE2 in transgenic P. ginseng was performed by in situ hybridization. PgSQE1 gene regulates saponin biosynthesis due to the lowered production of ginsenosides in PgSQE1RNAi lines. PgSQE2 gene positively regulates sterol synthesis [79]. The characterization of the transcriptome of Panax notoginseng root has been made using NGS technology based on 454 GSFLX Titanium platform. The dammaranediol synthase (DS) is involved in the biosynthesis of dammarane-type saponins in P. notoginseng. The candidate cytochrome P450 (Pn02132 and Pn00158) UDP-glycosyltransferase (Pn00082) gene catalyzed the conversion of dammaranediol-II or β -amyrin to various ginsenosides and the modification of ginsenosides. The identification of SSRs genes provided gene discovery, transcriptional regulation and marker-assisted breeding of drug resources by the EST analysis [80].
Considerable effort has recently been made in the plant cell culture of valuable medicinal plants for large scale production of secondary plant metabolites economically. The novel method for in vitro multiplication of Bacopa monniera rooting through cell culture has been studied in liquid media. The shoot tips were cultured on Murashige Skoog (MS) liquid medium containing 1.0 mg/l benzylaminopurine (BAP) and 0.5 mg/l kinetin (Kn) at pH 5.8. The maximum shoot proliferation observed was 100%. The root from seedling calli was cultivated in solid and liquid MS medium containing 0.5 mg/1 1-naphthalene acetic acid (NAA). The maximum rooting (100%) was obtained by 3 weeks in liquid medium. This might be due to better uptake of nutrients as the larger surface areas of roots were in contact with the liquid medium and also because the liquid medium helps in maintaining O2:CO2 balance [81]. The shoots regenerated on Murashige Skoog (MS) medium with 2 mgl-1 kinetin (Kn), pH set at 4.5, with 2% sucrose accumulated highest content of bacoside A (13.09 mg g-1DW) of B. monniera. This might be due to stressful condition of inadequate supply of carbon to the cultures. Accumulation of regenerated shoot biomass was highest on MS medium with 2% sucrose at pH 5.8 [82].
The genetic transformation of Bacopa monniera for large scale production of bacopasaponins has been developed by the Ri-T DNA of two wild type strains LBA9402 and A4 of Agrobacterium rhizogenes. The axenic shoots were cultured on Murashige and Skoog’s basal medium (MSO) with 3% sucrose and 0.8% agar for 2 weeks. The two strains A4 and LBA9402 were grown in 10 ml YMB liquid medium containing 10 mM acetosyringone at pH 7.0 and 28°C for 20-24 h in dark. The axenic transformed root cultures were established on 20 ml MSO medium containing 500 mgl-1 ampicillin for four weeks with A. rhizogenes LBA9402 and A4 separately. The transformed callus and shoot cultures were established on solid MSO with ampicillin 500 mgl-1 from transformed roots with A. rhizogenes LBA9402 and A4 for four weeks. The PCR and RT-PCR analysis confirmed the rolA, rolB and rolC genes of Ri-TR-DNA involved in LBA-9402 transformed cultures at the transcription level. Ri-transformed callus line wc2 showed significantly higher biomass accumulation as compared to callus line wc1 after 35 days of culture on MSO medium. In A4 transformed plants, the four bacopasaponins bacopasaponin D, bacopasaponin F, bacopaside II and bacopaside V were enhanced five times than TW TC plants. Bacopasaponin D was the principal bacopasaponin accumulated maximally (0.86 ± 0.02%) and enhanced 2.25 fold than WT TC plants. Bacopaside V accumulated five times (0.06 ± 0.001%) than WT TC plants. Bacopasaponin E was the principal bacopasaponin accumulated maximally (0.72 ± 0.04%) in WT TC plants. The contents of bacoside A3 and bacopasaponin C in transformed plants were comparable (0.1 ± 0.03 and 0.35 ± 0.04%) with that of WT TC plants (0.08 ± 0.04 and 0.3 ± 0.03%) [83]. The effects of cryptogenin gene on saponin production of B. monniera in transgenic plants utilizing both Ri and Ti plasmids have been studied. Agrobacterium rhizogenes LBA9402 crypt (pRi1855+pBin19+crypt) and a disarmed A. tumefaciens strain LBA4404 crypt (pTi4404+pBin19+crypt) under the control of 35S CaMV viral promoter and the nos terminator were grown overnight in liquid YMB medium at pH 7.0. The roots redifferentiating from seedling calli transformed by Ri- T-DNa plus crypt were cultured on MS medium with reduced nitrogen strength, macronutrients, micronutrients, myoinositol, ammonium iron citrate, Gamberg’s vitamin, 3% w/v sucrose, and kanamycin 100 mgl-1, and then subcultured at 6-8 week interval. The transformed plants by Ri-T-DNA and the crypt gene were established on MS medium. The transformed plants were established by Ti-DNA plus crypt from microshoots, cultured on MS medium with cefotaxime (500 mgl-1) and kanamycin (100 mgl-1). The PCR and RT-PCR analysis of Ti crypt plants showed the presence and expression of the crypt gene. The Ri crypt transformed plants showed more than two fold enhancement of shoot biomass than root biomass. The bacopasaponins bacoside A3, bacopasaponin C, bacopasaponin D, bacopasaponin F, bacopaside II and bacopaside III were enhanced six fold in Ti crypt-transformed plants. Bacopasaponin D was maximally enhanced. Bacoside A3 was enhanced four times in Ri crypt plant than Ri transformed plant, while bacopaside II accumulated 3 times higher compared to Ri-transformed plants. Bacopaside III increased fourfold than Ri-transformed plants. Bacopaside V content in Ricrypt plants were recorded to be two fold increased from the Ri plants [84].
An efficient regeneration system has been developed via culture, callus induction, regeneration of callus with somatic embryos, somatic embryo formation, somatic embryo maturation, somatic embryo germination, multiplication and rooting of plants, multiple shoot bud induction from shoot crown explants and leaves in Chlorophytum borivillianum and standardization of optimum protocol for rapid mass scale propagation. The shoot crown bud of C. borivilianum were cultured on modified solid MS medium with 2-4 D (0.1 mg/l) and 0.2 mg/l kn. The somatic embryos formed, induced on 2-4 D (1.25 mg/l) and BAP (0.1 mg/l), were subcultured on the same medium. Modified MS medium with 0.2 mg/1 BAP and 0.01 mg/l NAA was more effective for shoot regeneration of embryogenic calli. The maximum root formation (95%) was observed in ½ strength MS medium containing 1 mg/1 IBA and 30 g/1 sucrose for 3-4 weeks. High multiplication efficiency, phenotypic stability and yield confirmed the efficacy of the protocol developed for the production of medicinal herb safed musli [85]. The enhancement of concentration in the tubers of safed musli through mycorrhizal fungi inoculation has been studied. The roots of C. borivilianum were inoculated by mycorrizal fungi in a green house experiment for 270 days. The saponin concentration was enhanced by Glomus mossyee (65.2 to 157.0%) followed by Glomus intraradicis (61.5 to 125.2%) and Glomus fasciculatum (13.0 to 113.1%) as compared to control treatment under different crop age. The higher rate of accumulation as saponins in mycorrizal plants may be due to higher rate of photosynthesis. The more accumulation of saponins in plant growth may be due to restriction of shoot growth resulting in the development of stress condition in safed masli plant. The mycorrizal fungi inoculated safed musli have higher potential of transfer of carbon to the tuber [86-93].
Report of New Triterpenoid Saponins during 2007- 2012
New triterpenoid saponins isolated during the period along with their natural distribution, available physical data and spectral data for their characterization are presented in Table 7. Structures 4-220 are aglycones of the various saponins and structures 221- 224 are those of special saponins.
| Source | Saponin, mp, [α]D, | Structure spectra recorded | Ref |
|---|---|---|---|
| Acanthopanax koreanum (Araliaceae) |
Acankoreoside J 197-200, -38, IR, 1H, 13C, 2D, ESIMS | Aglycone (171) Rha-4Glc-6Glc(CO2H-28) |
[94] |
| Acankoreoside K 225-230, -50, IR, 1H,13C, 2D, ESIMS | Aglycone (172) Rha-4Glc-6Glc(CO2H-28) |
[94] | |
| Acankoreoside L, 238-241, -46, IR, 1H, 13C, 2D, ESIMS | Aglycone (174) Rha-4Glc-6Glc(CO2H-28) |
[94] | |
| Acankoreoside M, 1H, 13C, 2D, ESIMS | Aglycone (175), Glc(OH-3β), Rha-4Glc-6Glc(CO2H-28) |
[95] | |
| Acankoreoside N, 1H, 13C, 2D, ESIMS | Aglycone (176), Rha-4Glc-6Glc(CO2H-28) |
[95] | |
| Acankoreoside O, 1H, 13C, 2D, ESIMS | Aglycone (177), Rha-4Glc-6Glc(CO2H-28) |
[95] | |
| A.senticosus | Acanthopanaxoside E, -20.8, IR, 1H, 13C, 2D, FABMS | Echinocystic acid (10) GlcA(OH-3β), Glc(CO2H-28) |
[96] |
| Acanthophyllum elatius | Compound 1, -7.1, 1H, 13C, 2D, FABMS | Gypsogenic acid (11), Gal(CO2H-23) Glc-6Gal(CO2H-28) 3ÃÆââ¬âÃââ⬠Glc |
[97] |
| A.lilacinum | Compound 1, -7.1, 1H, 13C, 2D, FABMS | Gypsogenic acid (11), Gal(CO2H-23) Glc-6Gal(CO2H-28) 3ÃÆââ¬âÃââ⬠Glc |
[97] |
| A.sordidum (Caryophyllaceae) |
Compound 2 -15.6, 1H, 13C, 2D, FABMS | Glc-6Gal(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc Gypsogenic acid (11) Glc-6Gal(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[97] |
| Achlionice violaecuspidata (Holothurioidea) |
Achlioniceoside A1, 228-229, 1H, 13C, 2D, ESIMS | Aglycone (180) (SO3Na-6’) Glc-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 (MeO-3’)Glc-3(SO3Na-6’)Glc-4Quin Aglycone (182) |
[98] |
| Achlioniceoside A2, 229-230, 1H, 13C,1H, 13C, 2D, ESI-MS | (SO3Na-6’) Glc-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 (MeO-3’) Glc-3(SO3Na-6’)Glc-4Quin |
[98] | |
| Achlioniceoside A3 225-227, UV, IR, 1H, 13C, 2D, ESI-MS |
Aglycone (189) (SO3Na-6’) Glc-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 (MeO-3’)Glc-3(SO3Na-6’)Glc-4Quin |
[98] | |
| Aesculus glabra (Sapindaceae) |
Aesculioside G1 +2.3,1H,13C, 2D, ESIMS |
Aglycone (89) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] |
| Aesculioside G2 -1.8, 1H, 13C, 2D, ESIMS |
Aglycone (85) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G3 +7.4, 1H, 13C, 2D, ESIMS |
Aglycone (98) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G4 1.3, 1H, 13C, 2D, ESIMS |
Aglycone (87) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G5 8.0, 1H, 13C, 2D, ESIMS |
Aglycone (97) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G6 -21.6, 1H, 13C, 2D, ESIMS |
Aglycone (94) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G7 -18.2, 1H, 13C, 2D, ESIMS |
Aglycone (127) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G8 -7.9, 1H, 13C, 2D, ESIMS |
Aglycone (128) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G9 -15.2, 1H, 13C, 2D, ESIMS |
Aglycone (99) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G10 -15.3, 1H, 13C, 2D, ESIMS |
Aglycone (95) Ara(f)-3GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G11 -22.3, 1H, 13C, 2D, ESIMS |2 Gal |
Aglycone (99) Ara(f)-3(OMe-6’)GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G12 -19.5, 1H, 13C, 2D, ESIMS |
Aglycone (86) Ara(f)-3(OMe-6’)GlcA(OH-3β) |2 Gal |
[99] | |
| Aesculioside G13 -15.7, 1H, 13C, 2D, ESIMS |
Aglycone (133) Ara(f)-3GlcA(OH-3β) |2 Glc |
[99] | |
| Aesculioside G14 -14.5, 1H, 13C, 2D, ESIMS |
Aglycone (83) Ara(f)-3GlcA(OH-3β) |2 Glc |
[99] | |
| Aesculioside G15 -26.9, 1H, 13C, 2D, ESIMS |
Aglycone (84) Ara(f)-3GlcA(OH-3β) |2 Glc |
[99] | |
| Aesculioside G16 -27.9, 1H, 13C, 2D, ESIMS |
Aglycone (90) Ara(f)-3GlcA(OH-3β) |2 Glc |
[99] | |
| A.pavia | Aesculioside IIe 1H, 13C, 2D, ESIMS |
Aglycone (132) Ara(f)-3GlcA(OH-3β) |2 Glc |
[100] |
| Aesculioside IIf 1H, 13C, 2D, ESIMS |
Aglycone (133) Ara(f)-3GlcA(OH-3β) |2 Glc |
[100] | |
| Aesculioside IIg 1H, 13C, 2D, ESIMS |
Aglycone (83) Ara(f)-3GlcA(OH-3β) |2 Gal |
[100] | |
| Aesculioside IIh 1H, 13C, 2D, ESIMS |
Aglycone (125) Ara(f)-3GlcA(OH-3β) |2 Glc |
[100] | |
| Aesculioside IIi 1H, 13C, 2D, ESIMS |
Aglycone (91) Ara(f)-3GlcA(OH-3β) |2 Glc |
[100] | |
| Aesculioside IIj 1H, 13C, 2D, ESIMS |
Aglycone (84) Ara(f)-3GlcA(OH-3β) |2 Gal |
[100] | |
| Aesculioside IIk 1H, 13C, 2D, ESIMS |
Aglycone (90) Ara(f)-3GlcA(OH-3β) |2 Gal |
[100] | |
| Aesculioside IIIa 1H, 13C, 2D, ESIMS |
Aglycone (85) Ara(f)-3GlcA(OH-3β) |2 Gal |
[100] | |
| Aesculioside IIIb 1H, 13C, 2D, ESIMS |
Aglycone (135) Ara(f)-3GlcA(OH-3β) |2 Gal |
[100] | |
| Aesculioside IIIc 1H, 13C, 2D, ESIMS |
Aglycone (134) Ara(f)-3GlcA(OH-3β) |2 Glc |
[100] | |
| Aesculioside IIId 1H, 13C, 2D, ESIMS |
Aglycone (96) Ara(f)-3GlcA(OH-3β) |2 Gal |
[100] | |
| Aesculioside IIIe 1H, 13C, 2D, ESIMS |
Aglycone (100) Ara(f)-3GlcA(OH-3β) |2 Glc |
[100] | |
| Aesculioside IIIf 1H, 13C, 2D, ESIMS |
Aglycone (88) Ara(f)-3GlcA(OH-3β) |2 Gal |
[100] | |
| Albizia chinensis (Leguminosae) |
Albizoside A, -34.4, UV, IR, & 13C, 2D, MALDITOFMS | Aglycone (101) Xyl-2Fuc-6Glc(OH-3β) [(2E,6S)-2-hydroxymethyl-6-methyl-2’,7’ octadienoyloxy-4’] Quin-4[(2’E,6’S)-2-ydroxy methyl-6-methyl-2’,7’-ctadienoyloxy]Quin- 4[(2’’E,6’’S)-2’,6′-dimethyl-2,7-ctadienoyloxy] Quin(OH-21β) Ara(f)-4Rha-2Glc(CO2H-28) |3 Glc |
[101] |
| Albizoside B -22.0, UV, IR, 1H MALDITOFMS |
Aglycone (101) Xyl-2Fuc-6Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Glc [(2E,6S)-2-hydroxymethyl-6-methyl-2’,7’-octadi enoyl-4’]Quin-4[(2’E,6’S)-2-hydroxymethyl-6-meth yl-2’,7’-octadienoyloxy]Quin-4[(2’’E,6’’S)-2’,6′-dime thyl-2,7-octadienoyloxy]Quin(OH-21β) Ara(f)-4Rha- 2Glc(CO2H-28) |3 Glc |
[101] | |
| Albizoside C -16.0, UV, IR, 1H & 13C, 2D, MALDITOFMS |
Aglycone (101) Xyl-2Ara-6Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Glc [(2E,6S)-2-hydroxymethyl-6-methyl-2’,7’-octadienoy -loxy]Quin-4[(2’E,6’S)-2’,6′-dimethyl-2,7- octadienoyloxy]Quin(OH-21β) Ara(f)-4Rha-2Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[102] | |
| Albizoside D -10.2, UV, IR, 1H & 13C, 2D, QFTMS |
Aglycone (101) Xyl-2Fuc-6Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Glc [(2E,6S)-2’,6′-dimethyl-2’,7’-octadienoyloxy]QuinAra(f)-4Rha-2Glc(CO2H-28) -4[(2’E,6’S)-2-hydroxymethyl-6-methyl-2,7-octadienoyloxy]Quin(OH-21β) ÃÆââ¬âÃâââ¬3 Glc |
[102] | |
| Albizoside E -14.3, UV, IR, 1H & 13C, 2D,QFTMS |
Aglycone (101) Xyl-2Ara-6Glc(OH-3β) [(2E,6S)-2-hydroxymethyl-6-methyl-2’,7’-octadien oyloxy-4’]Quin(2’E,6’S)-2-hydroxymethyl-6-methyl -2’,7’-octadienoyloxy]Quin-4[(2’’E,6’’S)-2’,6′-dimeth yl-2,7-octadienoyloxy]Quin(OH-21β) Ara-4Rha-2Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[103] | |
| A.coriaria | Coriarioside A -40.0, 1H & 13C, 2D,ESIMS,FABMS | Aglycone (106) Fuc-6Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Glc [(2E,6S)-2’,6′-dimethyl-6’-hydroxy-2’,7’-octadien- oyloxy]Quin-4[(2′E,6′S)-2’,6′-dimethyl-6’-hydroxy- 2’,7’-octadienoyloxy]Quin(OH-21β) Rha-2Glc(CO2H-28) |
[103] |
| Coriarioside B -30.0, 1H & 13C, |
[(2E,6S)-2-hydroxymethyl-6-methyl-2’,7’-octadi enoyloxy-4’](2’E,6’S)-2-hydroxymethyl-6-methyl- 2’,7’-octadienoyloxy]Quin-4[(2’’E,6’’S)-2’,6′-dimeth yl-2,7-octadienoyloxy]Quin(OH-21β) Fuc-6Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Glc Xyl-3Rha-2Glc(CO2H-28) |
[104] | |
| A.inundata | Compound1 +25, IR, 1H & 13C, 2D, ESIMS Compound 2 -6, IR, 1H & 13C, 2D, ESIMS |
Oleanolic acid (6) Ara-6(NHCOCH3-2’)Glc(OH-3β) |
[104] |
| Compound 2 -6, IR, 1H & 13C, 2D, ESIMS |
Acacic acid lactone (26) Ara-2Ara-6(NHCOCH3-2’)Glc(OH-3β) |
[105] | |
| A.procera | Saponin -0.3, 1H,13C,2D, FABMS |
Echinocystic acid (10) Xyl-2Ara-6(NHCOCH3-2’)Glc(OH-3β) |
[105] |
| Saponin +5.7, H, 13C, 2D, FABMS |
Echinocystic acid (10) Ara-2Fuc-6(NHCOCH3-2’)Glc(OH-3β) |
[105] | |
| Saponin -9.7, 1H, 13C, FABMS |
Echinocystic acid (10) Xyl-2Ara-6(NHCOCH3-2’)Glc(OH-3β) |
[106] | |
| Saponin 1 -15.1, 1H, 13C, |
Echinocystic acid (10) Xyl-3Gal-6(NHCOCH3-2’)Glc(OH-3β) Glc(OH-16α) |
[106] | |
| Saponin 2 1H, 13C, FABMS |
Echinocystic acid (10) Xyl-2Ara-6(NHCOCH3-2’)Glc(OH-3β) Glc(OH-16α) |
[106] | |
| Saponin 3 1H, 13C, FABMS |
Echinocystic acid (10) Ara-2Ara-6(NHCOCH3-2’)Glc(OH-3β) Glc(OH-16α) |
[106] | |
| Androsace umbellate (Primulaceae) |
Compound 1 -16.82, IR, 1H, 13C,2D, ESIMS | Aglycone(165) Xyl-2Glc-4Ara(OH-3β) |2 Glc |
[107] |
| Compound 2 -20.19, IR, 1H, 13C, |
Aglycone(165) Xyl-2Glc-4Ara(OH-3β) |
[107] | |
| Anemone hupehensis (Ranunculaceae) |
Compound 1 -33.6, IR, 1H,13C, (Ranunculaceae)ESIMS | Oleanolic acid (6) Rib-2Rha-2Ara(OH-3β) Glc-3Rha-4Glc-6Glc(CO2H-28) |
[108] |
| Compound 2 -20.2, IR, 1H,13C, ESIMS |
Oleanolic acid (6) Glc-4Ara(OH-3β) |2 Rib-3Rha |
[108] | |
| Compound 3 -29.3, IR, 1H,13C, ESIMS |
Hederagenin (8) Rib-3Rha-2Ara(OH-3β) Glc-2Rha-4Glc-6Glc(CO2H-28) |
[108] | |
| A.raddena | Raddeanoside R22 285-286, +18.8, IR, 1H, 13C, 2D, |
Oleanolic acid (6) Glc-4Ara(OH-3β) |2 Glc |
[109] |
| Raddeanoside R23 248-250, +124, IR, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc-4Ara(OH-3β) |2 Ara-3Rha |
[109] | |
| A.taipaiensis | Compound 3 IR, 1H,13C, ESIMS |
Oleanolic acid (6) Xyl-2Rha-2Ara(OH-3β) Glc(CO2H-28) |
[110] |
| Compound 1 236-239, +12.6, IR, 1H,13C, 2D, ESIMS |
Hederagenin (8) Glc-4Ara(OH-3β) |2 Xyl-3Rha |
[110] | |
| Compound 2 249-253, -5.1, IR, 1H,13C, 2D, ESIMS |
Oleanolic acid (6) Glc-4Glc-4Ara(OH-3β) |2 Xyl-3Rha |
[110] | |
| Antonea ovate (Loganaceae) |
Antonioside A -10.0,1H, 13C, 2D,ESIMS |
Aglycone (120) Xyl-2Glc-3GlcA(OH-3β) |2 Glc |
[111] |
| Antonioside B -7,1H, 13C, 2D, 2D, ESIMS |
Aglycone (121) Xyl-2Glc-3GlcA(OH-3β) |2 Glc |
[111] | |
| Antonioside C -11,1H, 13C, 2D, ESIMS |
Aglycone (122) Xyl-2Glc-3GlcA(OH-3β) |2 Glc |
[111] | |
| Antonioside D -13,1H, 13C, 2D, ESIMS |
Aglycone (53) Xyl-2Glc-3GlcA(OH-3β) |2 Glc |
[111] | |
| Antonioside E +15.7,1H, 13C, 2D, ESIMS |
Aglycone(96) Glc-4Ara-6Glc(OH-3β) |4|2 GlcGlc |
[112] | |
| Antonioside F +19.9,1H, 13C, 2D, ESIMS |
Aglycone (132) Glc-4Ara-6Glc(OH-3β) |3|2 GlcGlc |
[112] | |
| Antonioside G +18.2,1H, 13C, 2D, ESIMS |
Aglycone (123) Glc-4Ara-6Glc(OH-3β) |3|2 GlcGlc |
[112] | |
| Antonioside H +16.3, 1H, 13C, 2D, ESIMS |
Aglycone (124) Glc-4Ara-6Glc(OH-3β) |3|2 GlcGlc |
||
| Antonioside I +17.8,1H, 13C, 2D, ESIMS |
Aglycone (125) Glc-4Ara-6Glc(OH-3β) |3|2 GlcGlc |
||
| Antonioside J +18.1,1H, 13C, 2D, |
Aglycone (126) Glc-4Ara-6Glc(OH-3β) |3|2 GlcGlc |
||
| Aralia elata | Tarasaponin IV 215-220, -10.5, UV IR, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Ara(f)-4GlcA(OH-3β) |2 Glc |
[113] |
| (Araliaceae) | U Elatoside L 235-240, -18.2,V, IR, 1H, 13C, 2D,ESIMS |
Oleanolic acid (6) Ara(f)-4GlcA(OH-3β) |2 Glc-6Glc Glc-6Glc(CO2H-28) |
[113] |
| Ardisia gigantifolia (Myrsinaceae) |
Compound 1 -17.5,1H, 13C, 2D, ESIMS |
Aglycone (170) Rha-3Glc-4Ara(OH-3β) |2|2 Xyl Glc |
[114] |
| Compound 2 -9.5,1H, 13C, 2D, |
Aglycone (168) Rha-3Glc-4Ara(OH-3β) |2|2 Glc-3Xyl Glc |
[114] | |
| Comound 3 1H, 13C, 2D, ESIMS |
Aglycone (169) Rha-3Glc-4Ara(OH-3β) |2|2 Xyl Glc |
[114] | |
| A.pusilla | Triterpenoid saponin 3 -23.2,1H, 13C, 2D, ESITOFMS |
Aglycone (167) Glc-3Glc-3Glc-4Ara(OH-3β |2ÃÆââ¬âÃâââ¬2 Xyl Glc |
[115] |
| Triterpenoid saponin 4 +4.5,1H, 13C, 2D, ESITOFMS |
Aglycone (48) Rha-2Glc-4Ara(OH-3β) |2 Glc |
[115] | |
| Triterpenoid saponin 5 -11.3,1H, 13C, 2D, ESITOFMS |
Aglycone (40) Rha-2Glc-4Ara(OH-3β) |2 Glc |
[115] | |
| Arenaria montana (Caryophyllaceae) |
Compound 1 -21.2, IR, 1H, 13C | Echinocystic acid (10) Glc(OH-3β) Xyl-4Rha(CO2H-28) |2 Rha |
[116] |
| Compound 2 -17.3, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Glc(OH-3β) Ara-3Xyl-4Rha(CO2H-28) |2 Rha |
[116] | |
| Compound 3 -12.6, IR, 1H, 13C,2D, ESIMS Apio(f)- 3Xyl-4Rha |
Caulophyllogenin (16) Glc(OH-3β) Apio(f)- 3Rha(CO2H-28) |2 Apio(f)- 3Xyl-4Rha |
[116] | |
| Astragalus eremophilus (Leguminosae) |
EremophilosideA IR, 1H, 13C, 2D, MALDITOFMS |
(2) | [117] |
| MALDITOFMS Eremophiloside B 270-272, -2.5, IR, 1H, 13C, 2D, |
Aglycone (217) Rha(OH-3β) Rha(OH-6α) |
[117] | |
| MALDITOFMS Eremophiloside C 278-280, +23 IR, 1H, 13C, 2D, |
Xyl(OH-25) Aglycone (218) Ara-2Xyl(OH-3β) |
[117] | |
| MALDITOFMS Eremophiloside D 289-291, +10.3 IR, 1H, 13C, 2D, |
Aglycone (211) Ara-2Xyl(OH-3β) |
[117] | |
| MALDITOFMS Eremophiloside E 275-278, -9.9, IR, 1H, 13C, 2D, |
Aglycone (208) Ara-2Xyl(OH-3β) |
[117] | |
| MALDITOFMS Eremophiloside F 285-288, +11.1, IR, 1H, 13C, 2D, |
Aglycone (212) Ara-2Xyl(OH-3β) |
[117] | |
| MALDITOFMS Eremophiloside G 160-162, -30.8, IR, 1H, 13C, 2D, |
Aglycone (209) Ara-2Xyl(OH-3β) |
[117] | |
| Eremophiloside G 160-162, -30.8, IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (209) Ara-2Xyl(OH-3β) |
[117] | |
| Eremophiloside H IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (213) Ara-2Xyl(OH-3β) |
[117] | |
| Eremophiloside I 170-173, -12.0 IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (220) Ara-2Xyl(OH-3β) |
[117] | |
| Eremophiloside I 170-173, -12.0 IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (210) Ara-2Xyl(OH-3β) |
[117] | |
| Eremophiloside K 269-271, +7.5 IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (211) Ara-2Xyl(OH-3β) |
[117] | |
| A.tauricolus | Compound 2 +13.1, IR, 1H,13C, 2D MALDITOFMS |
Aglycone (55) Rha-2Xyl-2GlcA(OH-3β) |
[118] |
| Compound 3 +9.3, IR, 1H, 13C, 2D,MALDITOFMS |
Aglycone (57) Rha-2Glc- 2GlcA(OH-3β) Glc(OH-29) |
[118] | |
| Compound 4 +22.4, IR, 1H, 13C, 2D,MALDITOFMS |
Aglycone (56) Rha-2Xyl-2GlcA(OH-3β) Rha(OH-21β) |
[118] | |
| Compound 5 +21.1, IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (56) Rha-2Glc- 2GlcA(OH-3β) |
[118] | |
| Compound 6 +15.6, IR, 1H, 13C, 2D, Rha-2Glc- 2GlcA(OH-3β) MALDITOFMS |
Aglycone (55) Rha-2Xyl- 2GlcA(OH-3β) Glc(CO2H-29) |
[118] | |
| Compound 7 +19.6, IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (58) Rha-2Xyl- 2GlcA(OH-3β) Rha(OH-22β) |
[118] | |
| Compound 9 +9, IR, 1H, 13C, MALDITOFMS |
Aglycone (50) Rha-2Glc- 2GlcA(OH-3β) |
[118] | |
| Compound 9 +9, IR, 1H, 13C, MALDITOFMS |
Aglycone (50) Rha-2Glc- 2GlcA(OH-3β) |
[118] | |
| Compound 11 +16.8, IR, 1H, 13C, MALDITOFMS |
Aglycone (41) Rha-2Glc- 2GlcA(OH-3β) |
[118] | |
| Compound 12 +16.8, IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (55) Glc- 2GlcA(OH-3β) Glc(CO2H-29) |
[118] | |
| Compound 14 +11.5, IR, 1H, 13C, 2D, MALDITOFMS |
Aglycone (55) Xyl- 2GlcA(OH-3β) Glc(CO2H-29) |
[118] | |
| Bellis perenni (Asteraceae) |
Perennisoside I +16.1, IR, 1H, 13C, 2D, FABMS | Aglycone (42) Glc-3Glc(CO2H-28) |2 Rha |
[118] |
| Perennisoside II +19.5, IR, 1H, 13C, |
Aglycone (42) Gal-3Glc(CO2H-28) |2 Rha |
[118] | |
| Perennisoside III +1.7, IR, 1H, 13C, 2D, FABMS |
Bayogenin (18) Glc(OH-3β) Glc-3Glc(CO2H-28) |2 Rha |
[118] | |
| Perennisoside IV +4.6, IR, 1H, 13C, 2D, FABMS |
Bayogenin (18) Glc(OH-3β) Gal-3Glc(CO2H-28) |2 Rha |
[118] | |
| Perennisoside V +7.1, IR, 1H, 13C, 2D, FABMS |
Aglycone (42) Glc(OH-3β) Glc-3Glc(CO2H-28) |2 Rha |
[118] | |
| Perennisoside VI +8.8, IR, 1H, 13C, 2D, FABMS |
Aglycone (42) Glc(OH-3β) Gal-3Glc(CO2H-28) |2 Rha |
[118] | |
| Perennisoside VII +14.3, IR, 1H, 13C, 2D, FABMS |
Aglycone (42) Glc(OH-3β) Gal-3Glc(CO2H-28) |2 Rha |
[118] | |
| Perennisaponin A -7.8, IR, 1H, 13C, 2D, FABMS |
Polygalacic acid (24) Rha-3Xyl-4(OAc-2’)Rha-2(OAc-4’)Fuc(CO2H-28) |
[118] | |
| Perennisaponin B -13.4, 1H, 13C, 2D, ESIMS |
Polygalacic acid Rha-3Xyl-4(OAc-2’)Rha-2[Methyl(S)- (+)-3-hydroxybutryloyloxy-4’]Fuc(CO2H-28) |
[118] | |
| Perennisaponin C -13.3, 1H, 13C, 2D, ESIMS | Polygalacic acid (24) Rha(OH-3β) Xyl-4(OAc-2’)Rha-2[{Methyl(S)-(+)-3- hydroxybutryloyloxy}-4’]Fuc(CO2H-28) |
[118] | |
| Perennisaponin D -14.9, 1H, 13C, 2D, FABMS |
Polygalacic acid (24) Rha(OH-3β) Rha-3Xyl-4(OAc-2’)Rha-2[[{Methyl(S)-(+)-3-hydroxybutryloyloxy}-4’]Fuc(CO2H-28) |
[118] | |
| Perennisaponin E -13.7, 1H, 13C, 2D, ESIMS |
Polygalacic acid (24) Rha(OH-3β) Rha-3Xyl-4(OAc-2’)Rha-2[(S)-3- hydroxybutryloyloxy-4’]Fuc(CO2H-28) |
[118] | |
| Perennisaponin F -21.2, 1H, 13C, 2D, ESIMS |
Polygalacic acid (24) Rha(OH-3β) Rha-3Xyl-4(OAc-2’)Rha-2[{Methyl(S)- (+)-3-hydroxybutryloyloxy}-4’]Fuc(CO2H-28) |
[118] | |
| B. sylvestris | Besylvoside I -12.0,1H, 13C, 2D, ESIMS | Bayogenin (18) Xyl-6Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 |2 GlcGlc |
[119] |
| Besylvoside II +32.2, 1H, 13C, |
Bayogenin (18) Xyl-6Glc(CO2H-28) |
[119] | |
| Besylvoside III +3.0, 1H, 13C, 2D, ESIMS |
Bayogenin (18) Xyl-6Glc(CO2H-28) |
[119] | |
| Besylvoside IV -8.3, 1H, 13C, 2D, ESIMS |
Bayogenin (18) Glc-3Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Glc |
[119] | |
| Besylvoside V -4.3, 1H, 13C,2D, |
Bayogenin (18) Glc-2Glc(CO2H-28) |
[119] | |
| Besylvoside VI -6.8, 1H, 13C, 2D, ESIMS |
Bayogenin (18) Glc(OH-3β) Xyl-6Glc(CO2H-28) |3|2 GlcGlc |
[119] | |
| Blighia sapida (Sapindaceae) |
Blighoside A IR, 1H, 13C, 2D, | Hederagenin (8) Ara-4(OAc-3’)Glc-3Rha-2Ara(OH-3β) |
[120] |
| Blighoside B IR, 1H, 13C, 2D, ESITOFMS |
Oleanolic acid (6) Ara-4(OAc-3’)Glc-3Rha-2Ara(OH-3β) |
[120] | |
| Blighoside C IR, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) (OAc-4’,6’)Glc-3Rha-4Glc(OAc-3’,6’)- 3Rha-2Xyl-2Xyl(OH-3β) |
[120] | |
| Bohadschia marmorata (Holothuridae) |
Marmoratoside A IR, 1H, 13C, 2D, ESIMS | Aglycone (181) (OMe-3’)Glc-3Glc-4Xyl(OH-3β) |2 (OMe-3’)Glc-3Glc-4Quin |
[121] |
| 17α-hydroxy impatien- side A 209-211, -11, ESIMS |
Aglycone (184) (OMe-3’)Glc-3Glc-4Xyl(OH-3β) |2 (OMe-3’)Glc-3Glc-4Quin |
[121] | |
| Marmoratoside B 215-217, IR, 1H, 13C, 2D, ESIMS |
Aglycone (182) (OMe-3’)Glc-3Glc-4Xyl(OH-3β) |2 (OMe-3’)Glc-3Glc-4Quin |
[121] | |
| 25-acetoxy bivittoside D 205-207, -9.1, IR, 1H, 13C, 2D, ESIMS |
Aglycone (187) (OMe-3’)Glc-3Glc-4Xyl(OH-3β) |2 (OMe-3’)Glc-3Glc-4Quin |
[121] | |
| 17-hydroxy fuscocin- eroside B 224-226, -0.2,IR, 1H,13C, 2D, FABMS |
Aglycone (191) (OMe-3’)Glc-3Glc-4Quin-2(4’-SO3Na)Xyl(OH-3β) |
[122] | |
| 25-hydroxy fuscocin- eroside B 219-221, -8.3,IR, 1H, 13C, 2D, FABMS |
Aglycone (192) (OMe-3’)Glc-3Glc-4Quin-2(4’-SO3Na)Xyl(OH-3β) |
[122] | |
| Butyrospe- rmum parkii (Sapotacea) |
Parkioside A -7.6, IR, 1H, 13C, 2D, ESIMS Parkioside B -51.8, IR, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc(OH-3β) Xyl-3Rha-2Xyl(CO2H-28) Oleanolic acid (6) Apio(f)-3Glc(OH-3β) Apio(f)-3Xyl-4Rha-2Xyl(CO2H-28) ÃÆââ¬âÃâââ¬3 Rha |
[123] |
Camellia sinensis (Theaceae) |
Parkioside C -39.9, IR, 1H, 13C, 2D, ESIMS Theasaponin A6 1H,13C, 2D, FABMS |
Aglycone (92) Glc-2Ara-3GlcA(OH-3β) |2 Gal Aglycone (93) |
[123] |
| Theasaponin A7 1H,13C, 2D, FABMS Glc-2Ara-3GlcA(OH-3β) |
Aglycone (93) 3GlcA(OH-3β) |2 Gal |
[123] | |
| Theasaponin B5 1H,13C, 2D, FABMS |
Aglycone (130) Xyl-2Ara-3GlcA(OH-3β) |2 Gal |
[124] | |
| Catunaregan spinosa (Rubiaceae) |
Catunaroside A+10.4, IR, 1H, | Siaresinolic acid (25) Glc-3Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Xyl |
[124] |
| Catunaroside B +32.4, IR, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc-3Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Rha |
[124] | |
| Catunaroside C +37.5, IR, 1H, 13C, |
Siaresinolic acid (25) Glc-3Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Glc(CO2H-28) |
[125] | |
| Catunaroside D +68.5, IR, 1H, 13C, 2D, ESIMS |
Siaresinolic acid (25) Glc-3Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Xyl Glc(CO2H-28) |
[125] | |
| Cephalaria scoparia (Dipsacaceae) |
Scoposide A -3.3, IR, 1H, 13C 2D,FABMS |
Oleanolic acid (6) Ara(OH-3β) Glc-6Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Rha |
[60] |
| Scoposide B -4.5, IR, 1H, 13C, 2D, FABMS |
Oleanolic acid (6) Xyl-4Xyl-3Rha-4Glc(OH-3β) Glc-6Glc(CO2H-28) |
[60] | |
| Scoposide C -2.8, IR, 1H, 13C, 2D, FABMS |
Oleanolic acid (6) Glc-4Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Xyl-3Rha Glc-6Glc(CO2H-28) |
[60] | |
| Scoposide D -2.4, IR, 1H, 13C, 2D, FABMS |
Oleanolic acid (6) Xyl-3Rha-3Ara(OH-3β) Glc(CO2H-28) |
[60] | |
| Scoposide E -4.0, IR, 1H, 13C, 2D, FABMS |
Oleanolic acid (6) Xyl-3Rha-3Ara(OH-3β) |
[60] | |
| Clematis chinensis (Ranunculaceae) |
Clematochinenoside A -62.0, UV, IR, 1H, | Hederagenin (8) Glc-2Rha-6Glc-3(isoferuloyloxy-2’)Glc-4Glc-4Rib -3Rha-2Ara(OH-3β) |
[126] |
| Clematochinenoside B -31.0, UV, IR, 1H, 13C, 2D, ESIMS |
Hederagenin (8) Glc-2Rha-6Glc-3Glc-4Glc-4Rib-3Rha2Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[126] | |
| Clematochinenoside C -58.0, UV, IR, 1H, 13C, |
Hederagenin (8) (isoferuloyloxy-2’)Glc-4Glc-4Rib-3Rha-2Ara(OH-3β) ÃÆââ¬âÃâââ¬3 Rha-6Glc ÃÆââ¬âÃâââ¬4 Glc |
[126] | |
| Clematochinenoside D Hederagenin (8) -52.0, UV, IR, 1H, 13C, 2D, ESIMS |
Rha-4Glc-6Glc(CO2H-28) (isoferuloyloxy-3’)Glc-4Glc-4Rib-3Rha-2Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[126] | |
| Clematochinenoside E -44.0, UV, IR, 1H, 13C, 2D, ESIMS |
Hederagenin (8) (isoferuloyloxy-6’)Glc-4Glc-4Rib-3Rha2Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[126] | |
| Clematochinenoside F -39.0, UV, IR, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) (isoferuloyloxy-6’)Glc-4Glc-4Rib-3Rha-2Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[126] | |
| Clematochinenoside G -66.0, UV, IR, 1H, 2D, ESIMS |
Hederagenin (8) Glc-3(isoferuloyloxy-2’)Glc-4Glc-4Rib-3Rha-2Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[126] | |
| Codonopsis lanceolata (Campanulaceae) |
Codonolaside III -56.6, IR, 1H, 13C, 2D, ESIMS | Echinocystic acid (10) Xyl-3GlcA(OH-3β) Glc-4Ara(CO2H-28) |2 Xyl-4Rha |
[127] |
| Combretum sundaicum (Holothuriaceae) |
Compound 3 +18.4, UV, IR,1H, 13C, 2D, ESIMS | Aglycone (35) (OAc-3’)Rha(OH-23) |
[128] |
| Compound 4 +29.9, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (35) Rha(OH-23) |
[128] | |
| Compound 5 +54.2, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (33) Rha(OH-1α) |
[128] | |
| Compound 6 +27.9, UV, IR,1H, 13C, 2D, ESIMS |
Aglycone (34) Rha(OH-1α) |
[128] | |
| Compound 8 +51.3, UV, IR, 1H, 13C, 2D, ESIMS | Aglycone (38) Rha(OH-1α) |
[128] | |
| Compound 9 +41.2, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (36) (OAc-4’)Rha(OH-23) |
[128] | |
| Compound 10 +31.2, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (37) OAc-2’,4’)Rha(OH-6α) |
[128] | |
| Cucumaria okhotensis (Holothuriaceae) |
Okhotoside B1 273-275 , -5.0,IR, 1H, 13C, 2D, ESIMS | Aglycone (178) (OMe-3’)Glc-3Glc-4Glc-2(SO3Na-4’)Xyl(OH-3β) |
[129] |
| Okhotoside B2 244 , -9.0, IR, 1H, 13C, 2D, FABMS |
Aglycone (178) (OMe-3’)Glc-3(SO3Na-6’)Glc-4Glc-2(SO3Na-4’) Xyl(OH-3β) |
[129] | |
| Okhotoside B3 260-263, -10.0, IR, 1H, 13C, 2D, FABMS |
Aglycone (178) (SO3Na-6’,OMe-3’)Glc-3(SO3Na-6’)Glc-4Glc- 2Xyl(OH-3β) |
[130] | |
| Cyclamen repandum (Primulaceae) |
Repandoside 1H, 13C, 2D, APITOFMS | Aglycone (164) Glc-4Glc-4Ara(OH-3β) |
[131] |
| Dianthus versicolor (Caryophylaceae) |
Dianversicoside A+19.6, IR, 1H, 13C, 2D, ESIMS | Aglycone (39) Glc(CO2H-23) Glc-2[(3S)-3-hydroxy-methylglutarooyloxy- 6’]Glc-6Glc(CO2H-28) |
[131] |
| DianversicosideB -12.3, IR, 1H, 13C, 2D, ESIMS |
Aglycone (39) Glc-2[(3S)-3-hydroxy-3- methylglutarooyloxy-6’] Glc-6Glc(CO2H-28) |
[131] | |
| Dianversicoside C +22.8, IR, 1H, 13C, Glc(CO2H-23) 2D, ESIMS |
Gypsogenic acid (11) Glc-2[(3S)-3-hydroxy-3-methylglutarooyloxy-6’] Glc-6Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[131] | |
| Dianversicoside D +8.4, IR, 1H, 13C, 2D, ESIMS |
Gypsogenic acid (11) Glc-2[(3S)-3-hydroxy-3- methylglutarooyloxy-6’]Glc-6Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[131] | |
| Dianversicoside E -16.8, IR, 1H, 13C, 2D, ESIMS |
Aglycone (39) Glc(CO2H-23) Glc-2[(3S)-3-hydroxy-3-methylglutarooyloxy]Glc-6Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[131] | |
| DianversicosideF -8.5, IR, 1H, 13C, 2D, ESIMS |
Aglycone (39) Glc(CO2H-23) Glc-2Glc-6Glc(CO2H-28) |
[131] | |
| DianversicosideG -6.0, IR, 1H, 13C, 2D, ESIMS |
Gypsogenic acid (11) Ara(OH-3β) Glc-2Glc-6Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[131] | |
| Dodonaea visicosa (Sapindaceae) |
Dodoneaside A -44.4, IR, 1H, 13C, 2D, FABMS |
Aglycone (112) Ara(f)-3GlcA(OH-3β) |2 Glc |
[132] |
| Dodoneaside B -80.0, IR, 1H, 13C, 2D, FABMS |
Aglycone (119) Ara(f)-3GlcA(OH-3β) |2 Glc |
[132] | |
| Echinopsis macrogona (Cactaceae) |
Pachanoside C1-14.7, IR, 1H, 13C,2D, FABMS | Aglycone (27) Xyl-2Glc-2GlcA(OH-3β) |
[8] |
| Pachanoside E1 205-208, -28.1 IR, 1H, 13C, 2D, FABMS |
Aglycone (28) Xyl-2Glc-2GlcA(OH-3β) |
[8] | |
| Pachanoside F 110.2, IR, 1H, 13C, 2D, FABMS |
Aglycone (29) Xyl-2Glc-2GlcA(OH-3β) |
[8] | |
| Pachanoside G1 -14.6, IR, 1H, 13C, 2D, FABMS |
Aglycone (30) Xyl-2Glc-2GlcA(OH-3β) |
[8] | |
| Bridgeside A1 -33.5, IR, 1H, 13C, 2D, FABMS |
Bridgesigenin A (43) Rha-2Glc-2GlcA(OH-3β) |
[8] | |
| Bridgeside C1 -33.7, IR, 1H, 13C, 2D, FABMS |
Bridgesigenin C (44) Xyl-2Glc-2GlcA(OH-3β) |
[8] | |
| Bridgeside C2 -44.6, IR, 1H,13C, 2D, FABMS |
Bridgesigenin C (44) Rha-2Glc-2 (Na+)GlcA(OH-3β) |
[8] | |
| Bridgeside D1 -17.9, IR, 1H, 13C, 2D, FABMS |
Bridgesigenin D (45) Xyl-2Glc-2GlcA(OH-3β) |
[8] | |
| Bridgeside D2 198-201, -34.5, IR, 1H, 13C,2D, FABMS |
Aglycone (46) Rha-2Glc-2GlcA(OH-3β) |
[8] | |
| Bridgeside E1 205-208, -28.1, IR, 1H, 13C,2D, FABMS |
Bridgesigenin E (47) Xyl-2Glc-2GlcA(OH-3β) |
[8] | |
| Bridgeside G1 -36.5, IR, 1H, 13C, 2D, FABMS |
Bridgesigenin E (47) Rha-2Glc-2GlcA(OH-3β) |
[8] | |
| Elaeocarpus hainanensis (Elaeocarpaceae) |
Compound 2 +39.3, IR, 1H, 13C, 2D, ESIMS | Aglycone (204) Glc(OH-2β) |
[133] |
| Compound 3 +63.7, IR, 1H, 13C, 2D, ESIMS |
Aglycone (205) Glc(OH-3β) |
[133] | |
| Entada rheedei (Mimosaceae) |
Rheediinoside A -27.1, IR, 1H, 13C, 2D, ESIMS | Entagenic acid (15) Xyl-3(OAc-6’)Glc-6(CH3CONH-2’)Glc(OH-3β) ÃÆââ¬âÃâââ¬3 Xyl Apio(f)-3Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl |
[134] |
| Rheediinoside B -28.5, 1H, 13C, 2D, ESIMS |
Entagenic acid (15) Xyl-3Glc-6(CH3CONH-2’)Glc(OH-3β) ÃÆââ¬âÃâââ¬3 Xyl Apio(f)-3Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl |
[134] | |
| Rheediinoside C -20.5, IR, 1H, 13C, |
Entagenic acid (15) Xyl-3(OAc-6’)Glc-6(CH3CONH-2’)Glc(OH-3β) ÃÆââ¬âÃâââ¬3 Xyl Xyl-3Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl |
[134] | |
| Rheediinoside D -24.0, IR, 1H, 13C, 2D, ESIMS |
Entagenic acid (15) Xyl-2Ara-6(CH3CONH-2’)Glc(OH-3β) Apio(f)-3Xyl-2(OAc-6’)Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl |
[134] | |
| Erylus formosus (Holothurioidea) |
Eryloside R1 1H, 13C, 2D, ESIMS | Aglycone (198) Gal-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[3] |
| Eryloside T1 1H, 13C, 2D, ESIMS |
Aglycone (198) Xyl-2Glc-4Gal-3Gal-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[3] | |
| Eryloside T2 1H, 13C, 2D, ESIMS |
Aglycone (198) Ara-2Glc-4Gal-3Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[3] | |
| Eryloside T3 1H, 13C, 2D, ESIMS |
Aglycone (198) Glc-2Glc-4Gal-3Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[3] | |
| Eryloside T4 1H, 13C, 2D, ESIMS |
Aglycone (196) Xyl-2Glc-4Gal-3Gal-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[3] | |
| Eryloside T5 1H, 13C, 2D, ESIMS |
Aglycone (196) Ara-2Glc-4Gal-3Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[3] | |
| Eryloside T6 1H, 13C, 2D, ESIMS |
Aglycone (196) Glc-2Glc-4Gal-3Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[3] | |
| Fadogia ancylantha (Rubiaceae) |
Compound 1 221, -3.8, 1H, 13C, 2D, ESIMS | Oleanolic acid (6) Glc(OH-3β) Rha-2Glc(CO2H-28) |
[135] |
| Compound 2 229, -7.3, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc(OH-3β) Apio(f)-2Glc(CO2H-28) |
[135] | |
| Compound 3 233, -3.5, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc(OH-3β) Glc-2Gal(CO2H-28) |
[135] | |
| Gleditsia capsica (Leguminosae) |
Caspicaoside A -33.8, 1H, 13C, 2D, FABMS | Echinocystic acid (10) Xyl-2Ara-6Glc(OH-3β) [(6S),(2E)-2,6-dimethyl-6-hydroxy-2,7-octadienoyloxy-2’]-Ara-2Rha ÃÆââ¬âÃâââ¬6 Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl-3Xyl-4Rha |
[136] |
| Caspicaoside B -29.1,1H, 13C, 2D, FABMS |
Echinocystic acid (10) Xyl-2Ara-6Glc(OH-3β) [(6S),(2E)-6-hydroxy-2,6-dimethyl-6-hydroxy-2,7- octadienoyloxy-2’]Ara-2[(6’S),(2’E)-2’,6’-dimethyl- 2’,7’-octadienoyloxy]Rha ÃÆââ¬âÃâââ¬6 Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl-3Xyl-4Rha |
[136] | |
| Caspicaoside C -18.4,1H, 13C, 2D, FABMS |
Echinocystic acid (10) Xyl-2Ara-6Glc(OH-3β) [(6S),(2E)-6’-hydroxy-2,6-dimethyl-2,7-octadienoyl oxy-2’]Ara-2[(6S),(2E)-6’-hydroxy-2’,6’-dimethyl- 2,7-octadienoyloxy-2’][(6’’S),(2’’E)-6’’-hydroxy- 2’’,6’’-dimethyl-2’’,7’’-octadienoyloxy]Rha ÃÆââ¬âÃâââ¬6 Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl-3Xyl-4Rha |
[136] | |
| Caspicaoside D -35.4,1H, 13C, 2D, |
Echinocystic acid (10) Xyl-2Ara-6Glc(OH-3β) [(6S),(2E)-6-hydroxy-2,6-dimethyl-2,6- dimethyl-2,7-octadienoyloxy-2’]Rha ÃÆââ¬âÃâââ¬6 Glc(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl-3Xyl-4Rha |
[136] | |
| Gordonia chrysandra (Theaceae) |
Gordonoside A -3.5, UV, IR, 1H, 13C, 2D, ESIMS | Aglycone (131) Ara-3GlcA(OH-3β) |
[137] |
| Gordonoside B -3.5, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (114) Ara-3GlcA(OH-3β) |
[137] | |
| Gordonoside C -5.9, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (136) Ara-3GlcA(OH-3β) |
[137] | |
| Gordonoside D -23.0, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (137) Ara-3GlcA(OH-3β) |
[137] | |
| Gordonoside E +1.2, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (115) Ara-3GlcA(OH-3β) |
[137] | |
| Gordonoside F -5.9, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (116) Ara-3GlcA(OH-3β) |
[137] | |
| Gordonoside G -1.0, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (113) Ara-3GlcA(OH-3β) |
[137] | |
| Gordonoside H -2.0, UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (117) Ara-3GlcA(OH-3β) |
[137] | |
| Gordonoside I 241-242, -2.7,UV, IR, 1H, 13C, 2D, ESIMS |
Aglycone (118) Xyl-2Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[138] | |
| Gordonoside J 239-240, 4.8,UV, IR, 1H, 13C,2D, ESIMS |
Aglycone (129) Xyl-2Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Gal |
[138] | |
| Gordonoside K 226-227, -7.0,UV, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Ara-3GlcA(OH-3β) Glc(CO2H-28) |
[138] | |
| Gordonoside L 234-235, UV, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Ara-2Ara-3GlcA(OH-3β) Glc(CO2H-28) |
[138] | |
| Gordonoside M 235-236, UV, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Rha-2Ara-3GlcA(OH-3β) Glc(CO2H-28) |
[138] | |
| Gordonoside N 238-239, -4.8,UV, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Xyl-2Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[138] | |
| Gordonoside O 232-233, -7.3,UV, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Xyl-2Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Gal Glc(CO2H-28) |
[138] | |
| Gordonoside P 227-228, -12.5,UV, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Xyl-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Rha-3Glc |
[138] | |
| Gypsophila aldhamiana (Caryophyllaceae) |
Gypsosaponin A -5.0, IR, 1H, 13C,2D, ESIMS | Quillaic acid (12) Xyl-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Gal Ara-2Ara-3Xyl-4Rha-2Fuc(CO2H-28) |
[139] |
| Gypsosaponin B -1.6, IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3(OMe-6’)GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Glc Xyl-4Rha-2Fuc(CO2H-28) ÃÆââ¬âÃâââ¬2 Glc |
[139] | |
| GypsosaponinC +14.9, IR, 1H, 13C, 2D, ESIMS |
Gypsogenic acid (11) Glc(CO2H-23) Glc-6Glc(CO2H-28) ÃÆââ¬âÃâââ¬3 Glc |
[140] | |
| G.perfoliata | Compound 1 +0.9, IR, 1H, 13C, 2D, | Quillaic acid (12) Xyl-3GlcA(OH-3β) |2 Gal (OAc-3’,4’)Quin-4Fuc(CO2H-28) |3 Xyl-3Xyl-3Xyl-4Rha |
[140] |
| Compound 2 +5.1, IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3GlcA(OH-3β) |2 Gal (OAc-3’,4’)Quin-4Fuc(CO2H-28) |2 Xyl-4Rha |
[140] | |
| Compound 3 -4.0, IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3GlcA(OH-3β) |2 Gal (OAc-3’,4’) Quin-4Fuc(CO2H-28) |2 Ara-3Xyl-4Rha |
[140] | |
| Compound 4 IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3GlcA(OH-3β) |2 Gal [4-O-trans-p-methoxy-cinnamoyloxy]Fuc(CO2H-28) |2 Xyl-4Rha ÃÆââ¬âÃâââ¬3 Glc |
[140] | |
| Compound 5 IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3GlcA(OH-3β) |2 Gal [4-O-cis-p-methoxy-cinnamoyloxy]Fuc(CO2H-28) |2 Xyl-4Rha ÃÆââ¬âÃâââ¬3 Glc |
[140] | |
| Compound 6 IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3(OMe-6’)GlcA(OH-3β) |2 Gal [4-O-trans-p-methoxy-cinnamoyloxy]Fuc(CO2H-28) |2 Xyl-4Rha ÃÆââ¬âÃâââ¬3 Glc |
[140] | |
| Compound 7 IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3(OMe-6’)GlcA(OH-3β) |2 Gal [4-O-cis-p-methoxy-cinnamoyloxy]Fuc(CO2H-28) |2 Xyl-4Rha ÃÆââ¬âÃâââ¬3 Glc |
[140] | |
| Compound 8 IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3(OMe-6’)GlcA(OH-3β) |2 Gal [3-O-trans-p-methoxy-cinnamoyloxy]Fuc(CO2H-28) |2 Xyl-4Rha ÃÆââ¬âÃâââ¬3 Glc |
[141] | |
| Compound 9 IR, 1H, 13C, 2D, ESIMS |
Gypsogenin (14) Xyl-3(OMe-6’)GlcA(OH-3β) |2 Gal [3-O-cis-p-methoxy-cinnamoyloxy]Fuc(CO2H-28) |2 Xyl-4Rha ÃÆââ¬âÃâââ¬3 Glc |
[141] | |
| Helianthus caulophyllogenin annus (Asteraceae) |
Caulophyllogenin 239-243, -48.2, IR, 1H, 13C, 2D, ESIMS | Helianthoside 4 (16) Xyl-4Glc(OH-3β) |3 Rha Glc-4Rha-2Xyl(CO2H-28) |
[142] |
| Helianthoside 5 222-227, -46.7,IR, ESIMS | Echinocystic acid (10) Xyl-4Glc(OH-3β) |3 Rha Rha-2Glc(CO2H-28) |
[142] | |
| Hemsleya chinensis (Cucurbitaceae) |
Xuedanglycoside A +108.4, IR, 1H, 13C, 2D, FABMSXuedanglycoside B +98.0, IR, 1H, 13C, 2D, FABMS |
Aglycone (206) Glc(OH-2β) Aglycone (207) Glc(OH-2β) |
[142] |
| Xuedanglycoside C +34.8, IR, 1H, 13C, 2D, FABMS | Oleanolic acid (6) Xyl-6Glc(CO2H-28) |
[142] | |
| Holothuria axiloga (Microthele) |
Arguside F 238-240, -1.2, 1H, 13C, 2D, ESIMS | Aglycone (194) Glc-4Xyl(OH-3β) |2 Glc(MeO-3’)-3Glc-4Quin |
[4] |
| Impatienside B 206-208, -7.8, 1H, 13C, 2D, ESIMS |
Glycone (195) Glc-4Xyl(OH-3β) |2 Glc(MeO-3’)-3Glc-4Quin |
[4] | |
| Pervicoside D 170-172, -4.0,1H, 13C, 2D, ESIMS |
Aglycone (183) Glc-4Xyl(OH-3β) |2 Glc(MeO-3’)-3Glc-4Quin |
[4] | |
| H.scabra | Scabraside A 234–236, −11.3,IR, ESIMS |
Aglycone (190) (OMe-3’)Glc-3Glc-4Quin-2(-SO3Na-4’)Xyl(OH-3β) |
[5] |
| Scabraside B ESIMS 236–238, −14.9,IR, |
Aglycone (193) (OMe-3’)Glc-3Glc-4Quin-2(-SO3Na-4’)Xyl(OH-3β) |
[5] | |
| Hydrocotyle bonariensis (Umbellferae) |
Bonarienoside A -26.4, 1H, 13C, 2D, ESIMS | Aglycone (102) Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[143] |
| Bonarienoside B -8.5, 1H, 13C, 2D, ESIMS |
Aglycone (103) Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[143] | |
| Bonarienoside C -13.0, 1H, 13C, 2D, ESIMS |
Aglycone (66) Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[143] | |
| Bonarienoside D -15.3, 1H, 13C, 2D, ESIMS |
Aglycone (67) Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[143] | |
| Bonarienoside E 1H, 13C, 2D, ESIMS |
Aglycone (104) Ara-3GlcA(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[143] | |
| H.sibthorpioides | Hydrocosisaponin A -1.6, IR, 1H, 13C, 2D, |
Aglycone (68) Ara-6Glc(OH-3β) |2 Glc |
[144] |
| Hydrocosisaponin B -2.7, IR, 1H, 13C, 2D, |
Aglycone (69) Ara-6Glc(OH-3β) |2 Glc |
[144] | |
| Hydrocosisaponin C -6.8, IR, 1H, 13C, |
Aglycone (70) Ara-6Glc(OH-3β) |2 Glc |
[144] | |
| Hydrocosisaponin D -2.8, IR, 1H, 13C, |
Aglycone (70) Ara-6Glc(OH-3β) ÃÆââ¬âÃâââ¬4 |2 GlcGlc |
[144] | |
| Hydrocosisaponin E -1.9, IR, 1H, 13C, 2D, |
Aglycone (71) Ara-6Glc(OH-3β) ÃÆââ¬âÃâââ¬4 |2 GlcGlc |
[144] | |
| Hydrocosisaponin F -4.1, IR, 1H, 13C, FABMS |
Aglycone (72) Ara-6Glc(OH-3β) ÃÆââ¬âÃâââ¬4 |2 GlcGlc |
[144] | |
| Ilex asprella (Aquifoliaceae) |
Asprellanoside A-2.15, IR, 1H, 13C, FABMS | Pomolic acid (142) (SO3H-3’)Xyl(OH-3β) Glc(CO2H-28) |
[145] |
| Asprellanoside B -3.42, IR, 1H, 13C, FABMS |
Ilexgenin B (158) (SO3H-3’)Xyl(OH-3β) |
[145] | |
| Ilexasoside A -2.15, IR, 1H, 13C, 2D, FABMS |
Pomolic acid (142) (SO3H-3’)Xyl(OH-3β) Glc(CO2H-28) |
[145] | |
| Asprellanoside B -3.42, IR, 1H, 13C, 2D, FABMS |
Ilexgenin B (158) (SO3H-3’)Xyl(OH-3β) |
[145] | |
| Ilexasoside A +10.3, IR, 1H, 2D, ESIMS |
Pomolic acid (142) (6’-OMe)GlcA(OH-3β) |
[145] | |
| IlexasosideB +2.8, IR, 1H, 13C, 2D, ESIMS |
Pomolic acid (142) (SO3Na-3’)(6’-OMe)GlcA(OH-3β) |
[145] | |
| Ilexasoside C -11.4,IR, 1H, 13C, 2D, 2D, ESIMS |
Pomolic acid (142) (6’-OMe)GlcA(OH-3β) Glc(CO2H-28) |
[145] | |
| Ilexasoside D -8.6, IR, 1H, 13C, 2D, ESIMS |
Pomolic acid (142) (SO3Na-3’)(6’-OMe)GlcA(OH-3β) Glc(CO2H-28) |
[146] | |
| Ilexasoside E 224-226,+104.5,IR, 1H, 13C, 2D, ESIMS |
Aglycone (151) (6’-OMe)GlcA(OH-3β) Glc(CO2H-28) |
[146] | |
| Ilexasoside F 213-215,+104.5,IR, 1H, 13C, 2D, ESIMS |
Aglycone (154) (6’-OMe)GlcA(OH-3β) Glc(CO2H-28) |
[146] | |
| Ilexasoside G 212-214, -29.5, IR, 2D,ESIMS |
Aglycone (157) (6’-OMe)GlcA(OH-3β) Glc(CO2H-28) |
[146] | |
| Ilexasoside H 222-223, -8.0, IR, 1H,13C,2D,ESIMS |
(223) | [146] | |
| Impatiens siculifer (Balsaminaceae) |
Impatienoside A -10.5, IR, 1H, 13C,2D,ESIMS | Aglycone (73) Gal-2GlcA(OH-3β) |
[146] |
| Impatienoside B +33.4, IR, 1H, 13C 2D, ESIMS |
Aglycone (54) GlcA(OH-3β) Glc-3Ara(OH-22β) |
[146] | |
| Impatienoside C +12.1, IR, 1H, 13C 2D, ESIMS |
Aglycone (54) Rha-2Ara-2GlcA(OH-3β) Glc-3Ara(OH-22β) |
[146] | |
| Impatienoside C +12.1, IR, 1H, 13C, 2D, ESIMS |
Aglycone (54) Rha-2Ara-2GlcA(OH-3β) Glc-3Ara(OH-22β) |
[147] | |
| Impatienoside D +12.0, IR, 1H, 13C, 2D, ESIMS |
Aglycone (54) Rha-2Gal-2GlcA(OH-3β) Glc-3Ara(OH-22β) |
[147] | |
| Impatienoside E +11.8, IR, 1H, 13C, 2D, ESIMS |
Aglycone (54) Glc-2Gal-2GlcA(OH-3β) Glc-3Ara(OH-22β) |
[147] | |
| Impatienoside F IR, 1H, 13C, 2D, ESIMS |
Aglycone (54) Xyl-2Gal-2GlcA(OH-3β) Glc-3Ara(OH-22β) |
[147] | |
| Impatienoside G -31.0, IR, 1H, 13C, 2D, ESIMS |
Echinocystic acid (10) GlcA(OH-3β) Xyl-4Rha-2Ara(CO2H-28) |
[147] | |
| Isolatocereus dumortieri Stenocereus alamosensis (Pereskioideae) |
Compound 1 | Dumortierigenin (23) Rha-2Glc-2(MeO-6’)GlcA(OH-3β) |
[23] |
| Compound 2 | (225) | [23] | |
| Compound 3 | (226) | [23] | |
| Compound 4 | Gummosogenin (22) Glc-3GlcA(OH-3β) |2 Glc |
[23] | |
| Compound 5 1H, 13C, 2D, FAMMS |
Gummosogenin (22) Glc-3(MeO-6’)GlcA(OH-3β) |2 Glc |
[23] | |
| Juglans sinensis (Juglanceae) |
Compound 3 +39.6,FABMS Compound 1 | Aglycone (143) Glc(CO2H-28) |
[148] |
| Compound 1 +39.8, 1H, 13C, 2D, FABMS |
Aglycone (75) Glc(CO2H-28) |
[149] | |
| Compound 3 +28.8, 1H, 13C, 2D, FABMS |
Aglycone (140) Glc(CO2H-28) |
[149] | |
| Compound 4 +28.9, 1H, 13C, 2D, FABMS |
Aglycone (144) Glc(CO2H-28) |
[149] | |
| Kalopanax pictus (Araliaceae) |
Triterpenoid saponin +55.3, IR, 1H, 13C, 2D, ESITOFMS | Aglycone (59) Glc(CO2H-28) |
[150] |
| Triterpenoid Saponin +11.0, IR,1H, 13C, 2D, ESITOFMS |
Hederagenin (8) (OAc-2’, 3’)Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[150] | |
| Triterpenoid Saponin +6.3, IR, 1H, 13C, 2D, ESITOFMS |
Hederagenin (8) (OAc-3’,4’)Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[150] | |
| Compound 1 -23.3, IR, 1H, 13C, 2D, ESITOFMS |
Aglycone (64) GlcA(OH-3β) |
[151] | |
| Compound 2 -12.8, IR, 1H, 13C, 2D, ESITOFMS |
Aglycone (80) GlcA(OH-3β) Glc(CO2H-28) |
[151] | |
| Compound 3 +4.5, IR, 1H, 13C, 2D, ESITOFMS |
Hederagenin (8) Gal-3Ara(OH-3β) Glc-6Glc(CO2H-28) |
[151] | |
| Lipastrotethya rhaphoxea (Dictyonellidae) |
Sarasinoside N 10.0, UV, IR, 1H, 13C, 2D, FABMS | Aglycone (199) (NHAc-2’)Gal-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 (NHAc-2’)Glc |
[152] |
| Sarasinoside O -2.6, UV, IR, 1H, 13C, 2D, FABMS |
Aglycone (202) (NHAc-2’)Gal-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 Glc-2Glc-6(NHAc-2’)Glc |
[152] | |
| Sarasinoside P -6.0, UV, IR, 1H, 13C, 2D, FABMS |
Aglycone (201) (NHAc-2’)Gal-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 Glc-2Glc-6(NHAc-2’)Glc |
[152] | |
| Sarasinoside Q -29.1, UV, IR, 1H, 13C, 2D, FABMS |
Aglycone (197) (NHAc-2’)Gal-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 Glc-2Glc-6(NHAc-2’)Glc |
[152] | |
| Sarasinoside R -12.4, UV, IR, 1H, 13C, 2D, FABMS |
Aglycone (203) (NHAc-2’)Gal-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 Glc-2Glc-6(NHAc-2’)Glc |
[152] | |
| Lysimachia clethroides (Primulaceae) |
Clethroidoside A +2.6, IR, 1H, 13C, 2D, ESI-TOFMS | Aglycone (48) Glc-4Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[153] |
| Clethroidoside B -37.2, IR, 1H, 13C, 2D, ESI-TOFMS |
Aglycone (72) ÃÆââ¬âÃâââ¬2 Glc |
[153] | |
| Clethroidoside C -30.5, IR, 1H, 13C, 2D, ESI-TOFMS |
Aglycone (166) Rha-2Glc-4Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[153] | |
| Clethroidoside D -55.4, IR, 1H, 13C, 2D, ESIMS |
Aglycone (166) Xyl-2Glc-4Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[153] | |
| Clethroidoside E -23.0, IR, 1H, 13C, 2D, ESI-TOFMS |
Aglycone (164) Rha-2Glc-4Ara(OH-3β) ÃÆââ¬âÃâââ¬2 (OAc-6’)Glc |
[153] | |
| Clethroidoside F -12.5, IR, 1H, 13C,2D, ESI-TOFMS |
Aglycone (163) Xyl-2Glc-4Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[153] | |
| Clethroidoside G -40.3, IR, 1H, 13C, 2D, ESI-TOFMS |
Aglycone (138) Glc-4Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[153] | |
| Clethroidoside H +30.8, IR, 1H, 13C, 2D, ESI-TOFMS |
Aglycone (141) Glc(OH-21β) Glc(OH-30) |
[153] | |
| L.heterogenea | Heterogenoside A IR, 1H, 13C, 2D, ESIMS |
Aglycone (62) Glc-2Ara(OH-3β) |
[154] |
| Heterogenoside B IR, 1H, 13C, 2D, ESIMS |
Aglycone (62) Xyl-2Glc-2Ara(OH-3β) |
[154] | |
| Heterogenoside C IR, 1H, 13C, 2D, ESIMS |
Aglycone (62) Xyl-2Glc-2Ara(OH-3β) |
[154] | |
| Heterogenoside D IR, 1H, 13C, 2D, ESIMS |
Aglycone (62) Xyl-2Glc-4Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Glc |
[154] | |
| Machilus yaoshansis (Lauraceae) |
Compound 1 -14.8, IR, 1H, 13C, 2D | Aglycone (160) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha |
[155] |
| Compound 2 +7.1, IR, 1H, 13C, 2D |
Aglycone (161) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha |
[155] | |
| Compound 3 +13.8, IR, 1H, 13C, 2D |
Aglycone (161) Xyl-3Glc(OH-3β) |
[155] | |
| Compound 8 +6.0, IR, 1H, 13C, 2D |
Aglycone (159) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha |
[155] | |
| Compound 9 -13.6, IR, 1H, 13C, 2D |
Aglycone (159) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha |
[155] | |
| Compound 10 +20.9, IR, 1H, 13C, 2D |
Aglycone (162) Xyl-3Ara(OH) |
[155] | |
| Compound 11 +2.2, IR, 1H, 13C, 2D |
Aglycone (162) Xyl-3Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Rha |
[155] | |
| Compound 12 -5.3, IR, 1H, 13C, 2D |
Aglycone (159) Xyl-3(OAc-6’)Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Rha |
[155] | |
| Compound 13 -9.3, IR, 1H, 13C, 2D |
Aglycone (159) Xyl-3(OAc-6’)Glc(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Glc-6Glc(OH-21β) |
[155] | |
| Medicago arabica (Fabaceae) |
Saponin 1 , 229-231, +37.5,1H, Queretaraic acid (9) 13C, 2D, ESIMS |
Aglycone (76) Ara-2GlcA(OH-3β) |
[156] |
| Saponin 2 232-233, +11.6, 1H, 13C, 2D, ESIMS |
Queretaraic acid (9) Glc(CO2H-28) Ara-2GlcA(OH-3β) Glc(CO2H-28) |
[156] | |
| Saponin 3 228-229,+33.3 1H, 13C, 2D, |
Aglycone (76) GlcA(OH-3β) Ara-2Glc(CO2H-28) |
[156] | |
| Saponin 4 231-233, +10.5, 1H, 13C, 2D, ESIMS |
Queretaraic acid (9) GlcA(OH-3β) Ara-2Glc(CO2H-28) |
[156] | |
| Saponin 5 214-215, +34.5, GlcA(OH-3β) |
Aglycone (76) Glc(CO2H-28) |
[156] | |
| Meryta denhamii (Araliaceae) |
Compound 1 +1.12, 1H, 13C, 2D, ESIMS | Aglycone (78) Rha-4Glc-6Glc(CO2H-28) |
[157] |
| Compound 2 +15.2, 1H, 13C, 2D, ESIMS |
Aglycone (78) Glc- 6Glc(CO2H-28) |
[157] | |
| Compound 3 +39.9,1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc-4Glc-3Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[157] | |
| Compound 4 +42.5,1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc-3Ara(OH-3β) |
[157] | |
| Compound 5 +23.6, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc-2GlcA(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[157] | |
| Compound 6 +4.3,1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Glc(CO2H-28) |
[157] | |
| Compound 7 +36.1,1H, 13C, 2D, ESIMS |
Echinocystic acid (10) Glc-4Glc-3Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[157] | |
| Compound 8 +45.1,1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc-4Glc-3Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[157] | |
| Nigella sativa (Ranunculaceae) |
Saponin 1 295, +21, IR, 1H, 13C, |
Aglycone (77) Xyl-2Rha-4Glc(OH-3β) |
[158] |
| Patrinia scabiosaefolia (Valerianaceae) |
Compound 1 -4.9, IR, 1H, 13C, 2D, ESIMS | (3) | [9] |
| Compound 3 -30.8, IR, 1H, 13C, 2D, ESIMS |
Aglycone (139) Xyl-2Glc(OH-3β) |
[9] | |
| Compound 4 -16.3, IR, 1H, 13C, 2D, ESIMS |
Oleanolic acid (6) Glc-4Xyl-2Rha-2Xyl(OH-3β) Glc(CO2H-28) |
[9] | |
| Pentacta quadrangularis (Cucumaridae) |
Pentactaside I 232-234, -17.4, 1H, 13C, 2D, ESIMS | Aglycone (186) Xyl-4Quin-2(NaSO3-4’)Xyl(OH-3β) |
[159] |
| Pentactaside II 230-233,-17.6, 1H, 13C, 2D, ESIMS |
Aglycone (178) Xyl-4Quin-2(NaSO3-4’)Xyl(OH-3β) |
[159] | |
| Pentactaside III 228-230, -36.1, 1H, 13C, 2D, ESIMS |
Aglycone (186) Quin-2(NaSO3-4’)Xyl(OH-3β) |
[159] | |
| Physena sessiliflora (Capparidaceae) |
Physenoside S1 +17.8, 1H, 13C, 2D, FABMS | Bayogenin (18) Glc(OH-3β) Ara-3Ara(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl-4Rha |
[6] |
| Physenoside S2 +18.8, 1H, 13C, 2D, FABMS |
Bayogenin (18) Glc(OH-3β) Apio(f)-3Ara(CO2H-28) ÃÆââ¬âÃâââ¬2 Xyl-4Rha |
[6] | |
| Physenoside S3 +12.7, 1H, 13C, 2D, FABMS |
Bayogenin (18) Glc(OH-3β) Rha-4Ara(CO2H-28) ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Xyl-4Rha Ara |
[6] | |
| Physenoside S4 +16.6,1H, 13C, 2D, FABMS |
Bayogenin (18) Glc -6Glc(OH-3β) Rha-4Ara(CO2H-28) ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Xyl-4Rha Ara |
[6] | |
| Physenoside S5 +1.3, 1H, 13C, 2D, FABMS |
Bayogenin (18) Glc(OH-3β) [{(8-oxo-16β-hydroxy)oxy}hexadecanoyloxy] Xyl-4Ara(CO2H-28) ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Xyl-4Rha Apio(f) |
[6] | |
| Physenoside S6 +10.2,1H, 13C, 2D, FABMS |
Bayogenin (18) Glc(OH-3β) [{(9-oxo-16β-hydroxy)} hexadecanoyloxy] Xyl-4Ara(CO2H-28 ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Xyl-4Rha Apio(f) |
[6] | |
| Physenoside S7 +5.2,1H, 13C, 2D FABMS |
Aglycone (32) Glc(OH-3β) [{(8-oxo-16β-hydroxy)} hexadecanoyloxy] Xyl-4Ara(CO2H-28) ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Xyl-4Rha Apio(f) |
[6] | |
| Physenoside S8 +1.2, 1H, 13C, 2D, FABMS |
Aglycone (32) Glc(OH-3β) [{(9-oxo-16β-hydroxy)} hexadecanoyloxy] Xyl-4Ara(CO2H-28) ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Xyl-4Rha Ara |
[6] | |
| Pithecellobium lucidum (Mimosaceae) |
Pithelucoside A -32.4, IR, 1H, 13C, 2D,FABMS | Aglycone (105) Xyl-2Fuc-Ara(f)-4Rha- |3 Quin |
[160] |
| Pithelucoside B –23.7, IR, 1H, 13C, 2D, FABMS |
Aglycone (106) Xyl-2Fuc-6Glc(OH-3β) Ara(f)-4Rha- |3 Glc |
[160] | |
| Pithelucoside C –13.5, IR, 1H, 13C, 2D, FABMS |
Aglycone (107) Xyl-2Fuc-6Glc(OH-3β) Ara(f)-4Rha-2Glc(CO2H-28) |3 Glc |
[160] | |
| Platycodon grandiÃÆïÃâìÃâââ¬Å¡orumon (Campanulaceae) |
Platyconic acid A -23.4, IR, 1H, 13C, 2D,MALDITOFMS | Aglycone (79) Glc-6Glc(OH-3β) Apio(f)-3Xyl-4Rha-2Ara(CO2H-28) |
[161] |
| Deapio-platyconic acid B Lactone -29.2, IR, 1H, 13C, 2D, ESIMS |
Aglycone (79) Glc-6Glc(OH-3β) Xyl-4Rha-2Ara(CO2H-28) |
[161] | |
| Deapio-platycodin D2 -14.4, IR, 1H, 13C, 2D, ESIMS |
Platycodigenin (7) Glc-3Glc(OH-3β)Xyl-4Rha-2Ara(CO2H-28) |
[161] | |
| Polygala tenuifolia (Polygalacea) |
Onjisaponin V +15.0, 1H, 13C, 2D | Presenegenin (13) Glc(OH-3β) (3’,4’,5’-trimethoxycinnamoyloxy-3’) Fuc(CO2H-28) ÃÆââ¬âÃâââ¬2 Gal-4Xyl-4Rha ÃÆââ¬âÃâââ¬3 (3-hydroxy-3-methylglutaroyloxy-5’)Apio(f) |
[162] |
| Onjisaponin W +25.0, 1H, 13C, 2D |
Presenegenin (13) Glc-(OH-3β) (3’,4’,5’-trimethoxycinnamoyloxy-3’)Fuc(CO2H-28) ÃÆââ¬âÃâââ¬2 Ara-3Xyl-4Rha ÃÆââ¬âÃâââ¬3 (3-hydroxy-3-methylglutaroyloxy-5’)Apio(f) |
[162] | |
| Onjisaponin X +4.0, 1H, 13C, 2D |
Presenegenin (13) Glc-(OH-3β) (3’,4’,5’-trimethoxycinnamoyloxy-4’)Fuc(CO2H-28) ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Ara-3Xyl-4Rha Gal ÃÆââ¬âÃâââ¬3 (3-hydroxy-3-methylglutaroyloxy-5’)Apio(f) |
[162] | |
| Onjisaponin Y -4.0, 1H, 13C, 2D |
Presenegenin (13) Glc-(OH-3β) (3’,4’,5’-trimethoxycinnamoyloxy-4’)Fuc(CO2H-28) ÃÆââ¬âÃâââ¬2 Gal-4Xyl-4Rha ÃÆââ¬âÃâââ¬3 (3-hydroxy-3-methylglutaroyloxy-5’)Apio(f) |
[162] | |
| Onjisaponin Z -5.0, 1H, 13C, 2D |
Presenegenin (13) Glc-(OH-3β) (p-trimethoxycinnamoyloxy-4’)Fuc(CO2H-28) ÃÆââ¬âÃâââ¬2 ÃÆââ¬âÃâââ¬3 Gal-4Xyl-4Rha Rha |
[162] | |
| Onjisaponin Vg -9.0, 1H, 13C, 2D |
Presenegenin (13) Glc-(OH-3β) Rha-3(3’,4’,5-triimethoxy-4’)Fuc(CO2H-28) 2ÃÆââ¬âÃââ⬠Xyl-4Rha |
[162] | |
| Rubus ellipticus (Rosaceae) |
Rubuside A +102.0, IR, 1H, 13C, 2D, FABMS | Aglycone (146) Glc(CO2H-28) |
[163] |
| Rubuside B +149.8, IR, 1H, 13C, 2D, FABMS |
Aglycone (147) Glc(CO2H-28) |
[163] | |
| Rubuside C +88.4, IR, 1H, 13C, 2D, FABMS |
Aglycone (149) Glc(CO2H-28) |
[163] | |
| Rubuside D +5.2, IR, 1H, 13C, 2D, FABMS |
Aglycone (155) Glc(CO2H-28) |
[163] | |
| Rubuside E +1.4, IR, 1H, 13C, 2D, FABMS |
Aglycone (156) Glc(CO2H-28) |
[163] | |
| Rubuside F +12.0, IR, 1H, 13C, 2D, FABMS |
Aglycone (149) Glc(CO2H-28) |
[163] | |
| Rubuside G -9.6, IR, 1H, 13C, 2D, FABMS |
Aglycone (153) Glc(CO2H-28) |
[163] | |
| Rubuside H -58.4, IR, 1H, 13C, 2D, |
Aglycone (152) Glc(CO2H-28) |
[163] | |
| Rubuside I -0.4, IR, 1H, 13C, 2D, FABMS |
Aglycone (173) Glc(CO2H-28) |
[163] | |
| Rubuside J -3.0, IR, 1H, 13C, 2D, FABMS |
Aglycone (150) Glc(CO2H-28) |
[163] | |
| Sapindus rarak (Sapindaceae) |
Rarasaponin I+16.8,1H, 13C, 2D, FABMS |
Hederagenin (8) (OAc-2’)Ara-3Rha-2Ara(OH-3β) |
[164] |
| Rarasaponin II +24.1, 1H, 13C, 2D, FABMS |
Hederagenin (8) (OAc-3’)Ara-3Rha-2Ara(OH-3β) |
[164] | |
| Rarasaponin III +31.1,1H, 13C, 2D, FABMS |
Hederagenin (8) (OAc-4’)Ara-3Rha-2Ara(OH-3β) |
[164] | |
| Rarasaponin IV +25.6,1H, 13C, 2D, FABMS |
Hederagenin (8) (OAc-3’, 4’)Ara-3Rha-2 (OAc-4’)Ara(OH-3β) |
[165] | |
| Rarasaponin V -8.3,1H, 13C, 2D, FABMS |
Hederagenin (8) (OAc-3’)Apio(f)-3Rha-2Ara(OH-3β) |
[165] | |
| Rarasaponin VI +1.9,1H, 13C, 2D, FABMS |
Hederagenin (8) (OAc-2’, 4’)Xyl-3Rha-2Ara(OH-3β) |
[164] | |
| Raraoside A +1.8, 1H, 13C, 2D, FABMS |
Aglycone (74) Rha-6Glc(OH-3β) |
[164] | |
| Sideroxylon foetidissimum (Sapotaceae) |
Saponin 1 1H, 13C, 2D, ESIMS | 16α-hydroxy protobassic acid (20) Glc-6Glc(OH-3β) Xyl-4Xyl-4Rha-2Ara(CO2H-28) ÃÆââ¬âÃâââ¬3 Rha |
[166] |
| Saponin 2 1H, 13C, 2D, ESIMS |
16α-hydroxy protobassic acid (20) Glc(OH-3β) Xyl-4Xyl-4Rha-2Ara(CO2H-28) ÃÆââ¬âÃâââ¬3 Rha |
[166] | |
| Saponin 3 1H, 13C, 2D, ESIMS |
16α-hydroxy protobassic acid (20) Glc-6Glc(OH-3β) Rha-3Xyl-4Rha-2Ara(CO2H-28) ÃÆââ¬âÃâââ¬3 Apio(f) |
[166] | |
| Saponin 5 1H, 13C, 2D, ESIMS |
16α-hydroxy protobassic acid (20) Glc(OH-3β) Rha-4Xyl-4Rha-2Ara(CO2H-28) ÃÆââ¬âÃâââ¬3 Apio(f) |
[166] | |
| Saponin 6 1H, 13C, 2D, ESIMS |
16α-hydroxy protobassic acid (20) Apio(f)-3Glc(OH-3β) Xyl-4Xyl-4Rha-2Ara(CO2H-28) ÃÆââ¬âÃâââ¬3 Rha |
[166] | |
| Stauntonia chinensis (Lardizabalaceae) |
Yemuoside YM17 IR, 1H, 13C, 2D, ESIITMS | Aglycone (80) Rha-4Glc-6Glc(CO2H-28) |
[22] |
| Yemuoside YM18, IR, 1H, 13C, 2D, ESITMS | Aglycone (82) Rha-4Glc-6Glc(CO2H-28) |
[22] | |
| Yemuoside YM19 IR, 1H, 13C, 2D, ESITMS |
Aglycone (81) Rha-4Glc-6Glc(CO2H-28) |
[22] | |
| Yemuoside YM20 IR, 1H, 13C, 2D, ESIITMS |
29-hydroxy oleanolic acid (26) Rha-4Glc-6Glc(CO2H-28) |
[22] | |
| Yemuoside YM26 +3.1, IR, 1H, 13C, 2D, ESIITMS |
Akebonic acid (17) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Rha-4Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM27 +7.9, IR, 1H, 13C, 2D, ESIITMS |
Akebonic acid (17) Gal-2Rha-2Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM28 +9.9, IR, 1H, 13C, 2D, ESIITMS |
Aglycone (51) Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Rha-4Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM29 +13.7, IR, 1H, 13C, 2D, ESIITMS |
Aglycone (51) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Rha-4Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM30 +17.4, IR, 1H, 13C, 2D, ESIITMS |
Aglycone (51) Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Glc-6Glc(CO2H-28) |
[167] | |
| YemuosideYM31 +10.2, IR, 1H, 13C, 2D, ESIITMS |
Aglycone (51) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM32 -6.6, IR, 1H, 13C, 2D, ESIITMS |
Hederagenin (8) Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Rha-4Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM33 -6.2, IR, 1H, 13C, 2D, ESIITMS |
Hederagenin (8) Xyl-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Rha-4Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM34 -2.0, IR, 1H, 13C, 2D, ESIITMS |
Hederagenin (8) Gal-2Rha-2Ara(OH-3β) Rha-4Glc-6Glc(CO2H-28) |
[167] | |
| Yemuoside YM35 2.7, IR, 1H, 13C, 2D, ESIITMS |
Hederagenin (8) Ara-3Ara(OH-3β) ÃÆââ¬âÃâââ¬2 Rha Glc-6Glc(CO2H-28) |
[167] | |
| Staurocucumis liouvillei (Cucumaridae) |
Liouvilloide A1, 228-230, -48, 1H, 13C, 2D, ESIFTMS | Aglycone (185) (OMe-3’)Glc-3(SO3Na-6’)Glc-4Quin-2(SO3Na-4’) Xyl(OH-3β) |
[20] |
| Liouvilloide A2 227-239, -53, 13C, 2D, ESIFTMS |
Aglycone (185) (OMe-3’)Quin-3(SO3Na-6’)Glc-4Quin-2(SO3Na-1H, 4’)Xyl(OH-3β) |
[20] | |
| Liouvilloide A3 225-228, 21, 1H, 13C, 2D, ESIFTMS |
Aglycone (186) (OMe-3’)Quin-3(SO3Na-6’)Glc-4Quin-2(SO3Na- 4’)Xyl(OH-3β) |
[20] | |
| Liouvilloide B1 239-241, -51, 1H, 13C, 2D, ESIFTMS |
Aglycone (185) (SO3Na-6’) (OMe-3’)Glc-3(SO3Na-6’)Glc-4Quin -2(SO3Na-4’)Xyl(OH-3β) |
[20] | |
| Liouvilloide B2 240-243, -18, 1H, 13C, 2D, ESIMS |
Aglycone (186) (SO3Na-6’) (OMe-3’)Glc-3(SO3Na-6’)Glc-4Quin- 2(SO3Na-4’)Xyl(OH-3β) |
[20] | |
| Sutherlandia frutescns (Fabaceae) |
Sutherlandioside A 210-220, +17.8, IR, 1H, 13C, 2D, ESIMS | Aglycone (200) Glc(OH-25) |
[168] |
| Sutherlandioside B 158, -26.8, IR, 1H, 13C, 2D, ESIMS |
Aglycone (214) Glc(OH-25) |
[168] | |
| Sutherlandioside C +15.0, IR, 1H, 13C, 2D, ESIMS |
Aglycone (215) Glc(OH-25) |
[168] | |
| Sutherlandioside D +58.0, IR, 1H, 13C 2D, ESIMS |
Aglycone (216) Glc(OH-25) |
[168] | |
| Synapta maculate (Holothurioidea) |
Synaptoside A -16, 1H, 13C, 2D, MALDITOFMS | Aglycone (188) (SO3Na-4’)Glc-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 (SO3Na-6’) (OMe-3’)GlcA-3Glc-4Quin |
[168] |
| Synaptoside A1 268-270, -3, 1H, 13C, 2D, MALDITOFMS |
Aglycone (179) (SO3Na-4’)Glc-4Xyl(OH-3β) ÃÆââ¬âÃâââ¬2 (SO3Na-6’) (OMe-3’)GlcA-3Glc-4Quin |
[168] | |
| Terminalia ivorensis (Combretaceae) |
Ivorenoside A -10.1,1H, 13C, 2D | (221) | [169] |
| Ivorenoside B -0.7, 1H, 13C, 2D |
(222) | [169] | |
| Ivorenoside C +13.3,1H, 13C, 2D |
Aglycone (63) Glc(CO2H-28) |
[170] | |
| T.superba Triterpenoid saponin 1 -13.2, IR, 1H, 13C, 2D, ESIMS |
Maslininc acid (21) Glc(CO2H-28) |
[170] | |
| Triterpenoid saponin 2 +4.2, IR, 1H, 13C, 2D, ESIMS |
Aglycone (31) Glc(CO2H-28) |
[171] | |
| Triterpenoid saponin 3 -18.1, IR, 1H, 13C, 2D, |
Aglycone (61) Glc(CO2H-28) |
[171] | |
| Triterpenoid saponin 4 -37.2, IR, 1H, 13C, 2D, |
Aglycone (49) Glc(CO2H-28) |
[171] | |
| T.tropophyll saniculagenin C | Terminaliaside A -6.1, 1H, 13C 2D, FABMS | 16-oxo-(angeloyloxy-3’)Glc(OH-3β) Glc-2Glc(CO2H-28) |
[172] |
| Verbesina virginica (Asteraceae) |
Verbesinoside A +36.2,UV, 1H, 13C, 2D, 2Glc(OH-3β) ESIMS |
Aglycone (111) Xyl-4Xyl-2Glc(OH-3β) |
[173] |
| Verbesinoside B +43.3, UV, 1H, 13C, 2D, ESIMS |
Aglycone (111) Xyl-4Glc(OH-3β) |
[173] | |
Verbesinoside C +22.5, UV, 1H, 13C, 2D, ESIMS |
Aglycone (108) Xyl-4Xyl-2Glc(OH-3β) |
[173] | |
| Verbesinoside D +27.7, UV, 1H, 13C, 2D, ESIMS |
Aglycone (109) Xyl-4Xyl-2Glc(OH-3β) |
[173] | |
| Verbesinoside E +31.9, UV, 1H, 13C, 2D, ESIMS |
Aglycone (110) Xyl-4Xyl-2Glc(OH-3β) |
[173] | |
| Verbesinoside F +24.2, UV, 1H, 13C, 2D, ESIMS |
Aglycone (52) Xyl-4Xyl-2Glc(OH-3β) |
[173] | |
| Xanthoceras sorbifolia (Sapindaceae) |
Sorbifoliaside A(4) 1H, 13C, 2D, ESIMS | Saniculagenin C Glc-6Glc(OH-2β) Glc-6Glc(OH-28β) |2 Glc |
[174] |
| Sorbifoliaside B saniculagenin C (5) 1H, 13C, 2D, ESIMS |
Glc-6Glc(OH-3β) Glc-6Glc(OH-28β) |2 Glc |
[174] | |
| Sorbifoliaside C Saniculagenin C (5) 1H, 13C, 2D, ESIMS |
Glc-6Glc(OH-3β) |2 Glc |
[174] | |
| Sorbifoliaside D 1H, 13C, 2D, ESIMS |
Saniculagenin C (4) Glc-6(angeloyloxy-3’)Glc(OH-3β) Glc-6Glc(OH-28β) |
[174] | |
| Sorbifoliaside E 1H, 13C, 2D, ESIMS |
16-oxo saniculagenin C (5) Glc-6(angeloyloxy-3’)Glc(OH-3β) Glc-6Glc(OH-28β) |
[174] | |
| Sorbifoliaside F 1H, 13C, 2D, ESIMS |
Saniculagenin C (4) Glc-6(angeloyloxy-3’)(OAc-4’)Glc(OH-3β) Glc-6Glc(OH-28β) |2 Glc |
[174] | |
| Bunkankasaponin F 1H, 13C, 2D,ESIMS |
Aglycone (65) Glc-2GlcA(OH-3β) (diangeloyloxy-3’,4’)Fuc(OH-21β) |
[174] | |
| Zygophyllum coccineum (Zygophyllacea) |
Zygophylloside S +0.050, 1H, 13C, ESIMS |
Quinovic acid (145) Ara-2Glc(OH-3β) |
[175] |
| Zygophylloside O +221-223, +23.7, 1H, 13C, ESIMS |
Quinovic acid (145) (2’-SO3H)Xyl(OH-3β) |
[175] | |
| Zygophylloside P +202-204, +20.0, 1H, 13C, ESIMS |
Quinovic acid (145) (2’-SO3H)Xyl(OH-3β) |
[175] |
Table 7: New Triterpenoid Saponins Isolated from 2007-2012.

(5) OH-3 β, 21 β, 22 α, 28, oxo-16, 16 oxo-saniculagenin C
(6) OH-3 β, CO2H-28, oleanolic acid
(7) OH-2 β, 3 β, 16 α, 23, 24, CO2H-28, platycodigenin
(8) OH-3 β, 23, CO2H –28, hederagenin
(9) OH-3 β, 30, CO2H-28, queretaroic acid
(10) OH-3 β, 16 α, CO2H-28, echinocystic acid
(11) OH-3 β, CO2H-23, 28, gypsogenic acid
(12) OH-3 β, 16 α, CHO-23, CO2H-28, quillaic acid
(13) OH-2 β, 3 β, 27, CO2H-23, 28, presenegenin
(14) OH-3 β, CHO-23, CO2H-28, gypsogenin
(15) OH-3 β, 15 α, 16 α, CO2H-28, entagenic acid
(16) OH-3 β, 16 α, 24, CO2H-28, caulophyllogenin
(17) OH -3 β, CO2H –28, 20:29-ene, 30-nor, akebonic acid
(18) OH- 2 β, 3 β, 23, CO2H-28, bayogenin
(19) OH- 3 β, 29, CO2H-28, 29-hydroxy oleanolic acid
(20) OH-2 β, 3 β, 6 β, 16 α, 23, CO2H-28, 16 α -hydroxy protobassic acid
(21) OH-2 α, 3 β, CO2H-28, maslinic acid
(22) OH- 3 β, 16 β, CHO-28, gummosogenin
(23) OH- 3 β, 22 α, 15 β :28 → lactone, dumortierigenin
(24) OH- OH - 2 β, 3 β, 16 α, 23, CO2H-28, polygalacic acid
(25) OH- 3 β, 19 α, CO2H-28, siaresinolic acid
(26) OH-3 β,16 α, 21 β :28 → lactone, acacic acid lactone
(27) OH-3 β, 30, OAc-21 β, 14 β :28 → lactone
(28) OH-3 β, 21 β, OAc-30, 14 β :28 → lactone
(29) OH-3 β, OAc-21 β, 30, 14 β :28 → lactone
(30) OH-3 β, OAc-21 β, 30, CO2H- 28, 13:14-ene
(31) OH-2 α, 3 β, 21 β, CO2H-28
(32) OH-2 β, 3 β, 16 α, CO2H-23, 28
(33) OH-1 α, OAc-3 β, 23, CO2H-29
(34) OH-1 α, 3 β, OAc- 23, CO2Me-29
(35) OH-1 α, 6 α, OAc-3 β, CO2H-29
(36) OH- 1 α, OAc-3 β, CO2Me-29
(37) OH-1 α, 3 β, CO2H-29
(38) OH-1 α, OAc-3 β, 23, CO2Me-29
(39) OH-3 β,16 α, CO2H- 23, 28
(40) OH-3 β, 28, 30
(41) OH-3 β, 21 α, 22 β, 23, 29
(42) OH-2 β, 3 β, OAc-23, CO2H-28
(43) OH-3 β, 21 β, 30, 15 β :28 → lactone bridgesigenin A
(44) OH-3 β, 30, OAc-21 β, 15 β :28 → lactone, bridgesigenin C
(45) OH-3 β, 21 β, OAc-30, 15 β :28 → lactone, bridgesigenin D
(46) OH-3 β, 21 β, OAc-30, 15 β :28 → lactone
(47) OH-3 β, OAc-21 β, 30, 15 β :28 → lactone, bridgesigenin E
(48) OH-3 β, 16 α, 28
(49) OH-2 α, 3 β, 23, 27, CO2H-28
(50) OH-3 β, 23, oxo-22, CO2H-29
(51) OH-3 β, 23, CO2H-28, 20:29-ene, 30-nor
(52) OH-3 β, CO2H-28, (E)-cinnamoyloxy-21 β, 15, 27-cyclo
(53) OH-3 β, 15 α, 16 α, 28, OAc-21 β, tigloyloxy-22 α
(54) OH-3 β, 21 β, 22 β, 24
(55) OH-3 β, 22 β, 23, CO2H-29
(56) OH-3 β, 21 β, 22 α, 23
(57) OH-3 β, 22 β, 23, 29
(58) OH-3 β, 22 β, 23
(59) OH-3 β, 6 β, 23, CO2H-28
(60) OH-3 β, 6 β, 16 α, CO2H-28
(61) OH-2 α, 3 β, 29, CO2H-28
(62) OH-3 β, 16 α, 23, 28
(63) OH-2 α, 3 β, 19 β, 23, oxo-11, CO2H-28
(64) OH-3 β, 6 β, 16 α, 23, CO2H-28
(65) OH-3 β, 16 α, 21 β, 28, OAc-22 α
(66) OH-3 β, 15 α, 16 α, 22 α, 28, OAc-21 β
(67) OH-3 β, 15 α, 16 α, 21 β, 22 α, 28
(68) OH-3 β, 16 α, 28, OAc-21 β, 22 α
(69) OH-3 β, 16 α, 21 β, OAc-22 α, 28
(70) OH-3 β, 16 α, 22 α, OAc-21 β, 28
(71) OH-3 β, 16 α, 22 α, 28, OAc-21 β
(72) OH-3 β, 16 α, 21 β, 22 α, OAc-28
(73) OH-3 β, 23, oxo-22
(74) OH-3 β, CHO-28
(75) OH-3 β, 23, oxo-1, CO2H-28
(76) OH-2 β, 3 β, 30, CO2H-28
(77) OH-3 β, 16 β, 23, OMe-11 α, OAc- 17 β
(78) OH-23, SO3H-3 β, CO2H- 28
(79) OH-3 β, 16 α, 23, CO2H- 28, 2 β :4 → lactone
(80) OH-3 β, 20 α, 30, CO2H- 28
(81) OH-3 β, 30, CO2H- 28, 20:21-ene
(82) OH-3 β, 20 β, 29, CO2H- 28
(83) OH-3 β, 15 α,16 α, 28, OAc- 22 α, angeloyloxy-21 β
(84) OH-3 β, 16 α, 28, OAc-22 α, tigloyloxy-21 β
(85) OH-3 β, 15 α,16 α, 22 α, OAc- 28, angeloyloxy-21 β
(86) OH-3 β, 16 α, 28, diangeloyloxy-21 β, 22 α
(87) OH-3 β, 16 α, 22 α, OAc-28, angeloyoxy-21 β
(88) OH-3 β, 15 α,16 α, 28, ditigloyloxy-21 β, 22 α
(89) OH-3 β, 15 α,16 α, 22 α, OAc-28, tigloyloxy-21 β
(90) OH-3 β, 16 α, 28, angeloyloxy-21 β, OAc- 22 α
(91) OH-3 β, 16 α, 23, 28, angeloyloxy-21 β, OAc- 22 α
(92) OH-3 β, 22 α, 23, 28, OAc-16 α, angeloyloxy-21 β
(93) OH-3 β, 23, 28, OAc-16 α, 22 α, angeloyloxy-21 β
(94) OH-3 β, 15 α, 16 α, 28, tigloyloxy-21 β, angeloyloxy-22 α
(95) OH-3 β, 16 α, 28, angeloyloxy-21 β, tigloyloxy- 22 α
(96) OH-3 β, 15 α, 16 α, 23, 28, ditigloyloxy- 21 β, 22 α
(97) OH-3 β, 15 α, 16 α, 22 α, 28, angeloyloxy- 21 β
(98) OH-3 β, 16 α, 22 α, OAc-28, tigloyloxy- 21 β
(99) OH-3 β, 16 α, 28, tigloyloxy- 21 β, angeloyloxy-22 α
(100) OH-3 β, 15 α, 16 α, 23, 28, tigloyloxy- 21 β, angeloyloxy-22 α
(101) OH-3 β, 16 α, CO2H –28, 21 β -[(2E,6S)-2-hydroxymethyl-6-methyl-6-hydroxyl}2,7- octadienoyloxy]acacic acid
(102) OH- 3 β, 15 α, 16 α, 28, OAc-22 α, 2-methylbutyroyloxy- 21 β
(103) OH- OH –3 β, 15 α, 16 α, 22 α, OAc- 28, 2-methylbutyroyloxy-21 β
(104) OH-3 β, 15 α, 16 α, 28, 2-methylbutyroyloxy-22 α
(105) OH-3 β, 16 α, CO2H-28, 21 β -[(6S)-hydroxy-(2E)-hydroxymethyl-6-methyl-2,7-octadienoyloxy]acacic acid
(106) OH-3 β, 16 α, CO2H-28, 21 β -[(6S)-hydroxy-(-(2E)- hydroxymethyl-6-methyl-6-hydroxy-2,7-octadienoyloxy]acacic acid
(107) OH-3 β, 16 α, CO2H-28, 21 β -[8-hydroxy-(2E)-hydroxymethyl-(6E)-methyl-2,7-octadienoyloxy]acacic acid
(108) OH-3 β, CO2H-28, p-methoxybenzoyloxy-21 β, 15, 27-cyclo
(109) OH-3 β, CO2H-28, (E)-trimethoxybenzoyloxy-21 β, 15, 27-cyclo
(110) OH-3 β, CO2H-28, (E)-p-methoxybenzoyloxy-21 β, 15,27-cyclo
(111) OH-3 β, CO2H-28, trimethoxybenzoyloxy-21 β, 15, 27-cyclo
(112) OH- 3 β, 15 α,16 α, 28, diangeloyloxy-21 β, 22 α
(113) OH-3 β, 15 α, 28, CO2Me-23, diangeloyloxy-21 β, 22 α
(114) OH-3 β, 23, 28, OAc-16 α, diangeloyloxy-21 β, 22 α
(115) OH-3 β, 28, CHO-23, diangeloyloxy-21 β, 22 α
(116) OH-3 β, 28, CO2Me-23, diangeloyloxy-21 β, 22 α
(117) OH-3 β, 15 α, 28, CO2Me-23, angeloyloxy-21 β,2-methylbutanoyloxy-22 α
(118) OH-3 β,16 α, 28, angeloyloxy-22 α
(119) OH-3 β,16 α, 28, angeloyloxy-22 α, epoxyangeloyloxy-21 β
(120) OH-3 β,16 α, 28, tigloyloxy- 22 α
(121) OH-3 β, 15 α, 16 α, 28, tigloyloxy- 22 α
(122) OH-3 β,16 α, 28, OAc-21 β, tigloyloxy- 22 α
(123) OH-3 β, 15 α, 16 α, 23, 28, tigloyloxy- 22 α, cinnamoyloxy-21β
(124) OH-3 β, 15 α, 16 α, 23, 28, OAc-22 α, cinnamoyloxy-21 β
(125) OH-3 β,16 α, 23, 28, OAc-22 α, tigloyloxy-21 β
(126) OH-3 β,16 α, 22 α, 23, OAc-28, tigloyloxy-21 β
(127) OH-3 β, 15 α, 16 α, 28, tigloyloxy-22 α, angeloyloxy-21 β
(128) OH-3 β, 16 α, 28, ditigloyloxy-21 β, 22 α
(129) OH-3 β, 15 α, 16 α, 28, angeloyloxy-22 α
(130) OH-3 β, 16 α, 22 α, angeloyloxy-21 β
(131) OH-3 β, 16 α, 23, 28, diangeloyloxy-21 β, 22 α
(132) OH-3 β, 15 α, 16 α, 23, 28, OAc-22 α, tigloyloxy- 21 β
(133) OH-3 β, 15 α, 16 α, 28, OAc-22 α, tigloyloxy- 21 β
(134) OH-3 β, 15 α, 16 α, 28, tigloyloxy- 21 β,2-methylpropanoyloxy-22 α
(135) OH-3 β, 15 α, 16 α, 24, 28, angeloyloxy- 21 β,2-methylpropanoyloxy-22 α
(136) OH-3 β, 28, diangeloyloxy- 21 β, 22 α
(137) OH-3 β, 28, OAc-16 α, 28, diangeloyloxy- 21 β, 22 α

(138) OH-3 β, 16 α, oxo-12, CHO-28
(139) OH-3 β, 12 β, 30, 28:13 β → γ-lactone

(140) OH-3 β, 23, CO2H-28
(141) OH-2 α, 3 β, 21 β, 30, 9:11-ene
(142) OH-3 β, 19 α, CO2H-28, pomolic acid
(143) OH-2 α, 3 α, 23, CO2H-28
(144) OH-3 β, 22 α, CO2H-28
(145) OH-3 β, CO2H, -27, 28, quinovic acid
(146) OH-2 α, 3 β, CO2H-28, 18:19-ene
(147) OH-2 α, 3 α, CO2H-28, 18:19-ene
(148) OH-2 α, 3 α, 23, CO2H-28, 18:19-ene
(149) OH-2 α, 3 β, CO2H-28, 19:29-ene
(150) OH-2 α, 3 α, 19 α, CHO-23, CO2H-28
(151) OH-3 β, CO2H-28, 18:19-ene

(152) OH-2 α, 3 β, 23, CO2H-28, 11:12, 13:18-ene
(153) OH-2 α, 3 α, CO2H-28, 11:12, 13:18-ene

(154) OH-3 β, CO2H-28, 18:19-ene, ilexolic acid
(155) OH-2 α, 3 β, CO2H-28, 19:20-ene
(156) OH-2 α, 3 α, CO2H-28, 19:20-ene
(157) OH-3 β, CO2H-28, 19:20-ene
(158) OH-3 β, 19 α, CO2H-28, ilexgenin B

(159) OH-3 β, 20(S), 21, 24:25-ene
(160) OH-3 β, 20(S), 23(R), 21:23 → lactone, 24:25-ene
(161) OH-3 β, 20(S), 23(R), 21:23 → lactone, 24:25-ene
(162) OH-3 β, 20(S), oxo-21, 24:25-ene

(163) OH- 3 β, 16 α, OAc-21 β, 22 α
(164) OH-3 β, 16 α
(165) OH-3 β, oxo-16, 30
(166) OH-3 β, oxo-16
(167) OH-3 β, 16 α, CHO-30
(168) OH-3 β, 16 α, OAc-30
(169) OH-3 β, 30, oxo-16
(170) OH-3 β, 16 α, 30

(171) OH-3 α, CO2H-23, 28, oxo-20
(172) OH-3 α, 20, 29, CO2H-23, 28
(173) OH-2 α, 3 β, 24, CO2H- 28, 20:29-ene
(174) OH-3 α, CO2H-23, 28, di-OMe-29
(175) OH-3 α, 11 α, 30, CO2H-23, 28, 20:29-ene
(176) OH-3 α, 11 α, 23, 30, CO2H-28, 20:29-ene
(177) OH-3 α, 19 β, CO2H-23, 28

(178) OH-3 β, OAc-16 β, 7:8, 25:26-ene
(179) OH-3 β, oxo-7, 23, 8:9-ene
(180) OH-3 β, 12 α, 24 α, 9:11, 25:26-ene
(181) OH-3 β, 12 α, 9:11, 25:26-ene
(182) OH-3 β, 12 α, 25, 9:11, 23:24-ene
(183) OH-3 β, 12 α, OAc-25, 9:11-ene
(184) OH-3 β, 12 α, 17 α, 9:11, 25:26-ene
(185) OH-3 β, oxo-16, 7:8, 25:26-ene
(186) OH-3 β, OAc-16 β, 7:8, 24:25-ene
(187) OH-3 β, 12 α, OAc-25, 9:11, 23:24-ene
(188) OH-3 β, oxo-23, 7:8-ene
(189) OH-3 β, 12 α, oxo-24, 9:11, 25:26-ene
(190) OH-3 β, 12 α, 17 α, 25:26-ene
(191) OH-3 β, 12 α, 17 α, oxo-22, 9:11-en
(192) OH-3 β, 12 α, OAc-16 β, 9:11, 24:25-ene
(193) OH-3 β, 9:11, 24:25-ene
(194) OH-3 β, 12 α, 25, oxo-22, 9:11-ene
(195) OH-3 β, 12 α, 17 α, oxo-22, 9:11-ene

(196) OH-3 β, CO2H-14, 8:9-ene, methylene-24
(197) OH-3 β, 9 α, oxo-23, 9:8-epoxy, 8:14, 24:25-ene
(198) OH-3 β, CO2H-14, 8:9, 24:25-ene
(199) OH-3 β, oxo-23, 8:9, 24:25-ene
(200) OH-1(S), 3(R), 24(S), 25, 7(S):10(S)-epoxy, 9,10-seco, 9:11-ene
(201) OH-3 β, 5 α, 9 β, oxo-23, 24:25-ene
(202) OH-3 β, 5 α, 9 α, oxo-23, 24:25-ene
(203) OH-3 β, 8 α, 9 α, 24:25-ene

(204) OH-2 β, 3 β, 20 β, oxo-11, 16 α :23 α –epoxy, 5:6, 24:25-ene
(205) OH-3 β, 20 β, 25, oxo-11, 16 α :23 α –epoxy, 5:6-ene
(206) OH-2 β, 3 α, 20, oxo-11, 16 α :23 α –epoxy, 5:6, 24:25-ene
(207) OH-2 β, 3 α, 16 α, 20, oxo-11, 22, 5:6, 25:26-ene
(208) OH-3 β, 6 α, 16 β :23 → lactone
(209) OH-3 β, 6 α, oxo-24 16 β :23:epoxy, 22:23-ene
(210) OH-3 β, 6 α, 23 α, 24 β, 16 β :23, 22 β :25-diepoxy
(211) OH-3 β, 23 α, 24 β, oxo-16, 24 β, 16 β :23, 22 β :25-diepoxy
(212) OH-3 β, oxo-6, 16 β :23 → lactone
(213) OH-3 β, 6 α, OMe-23 α, oxo-24, 16 β :23-epoxy
(214) OH--3(R), 7(S), 24(S), 25, oxo-1
(215) OH-3(R), 24(S), 25, oxo-1, 11
(216) OH-7(S), 24(S), 25, oxo-1, 2:3-ene
(217) OH-3 β, 6 α, 24(R), 25, OAc-16 β
(218) OH-3 β, 6 α, 16 β, 23(R), 24(R), 25
(219) OH-3 β, 16 β, 23(R), 24(R), 25, oxo-6
(220) OH-3 β, 6 α, OEt-23 α, oxo-24, 16 β :23-epoxy

(221) R=CH2OH
(222) R=COOH

Conclusions
The evaluation of structure-activity relationships has been increasing among the research activities of novel and potentially life-saving bioactive triterpenoid saponins. The study of triterpenoid saponins is being undertaken also to appraise saponin production through cell suspension culture system using metabolic engineering and post-genomics approaches for possible drug development and also for the development of varieties (green revolution) for future commercial exploitation of triterpenoid saponins economically. Herbal and marine drugs or medicines are gaining popularity in day-to-day life because of their low cost, wide occurrence in nature and least side effects. The natural triterpenoid saponins appear to be strong candidates for medicinal uses. The difficulties in isolation of natural saponins in considerable amounts due to their complex structural features has prompted global research communities to synthesize the designed compounds to gain access to adequate amounts of triterpenoid saponins in order to promote further pharmaceutical studies. So far, the inhibitors used in steel industry comprised synthetic compounds which are toxic and not eco-friendly whereas the inhibitors developed by studying the naturally occurring bioactive plant materials are biodegradable, nontoxic, eco-friendly. This may reduce the cost and environmental impact.
Acknowledgements
Financial support from CSIR, New Delhi is gratefully acknowledged.
References
- Garai S (2014)Triterpenoid saponins. Nat Prod Chem and Res 2: 1-13.
- Sticher O (2008)Natural Product isolation. Nat.Prod.Rep. 25: 517-554.
- Sugimoto S, Matsunami K, Otsuka H (2011) Medicinal Plants of Thailand. I Structures ofRheedeiosides A—D and cis-Entadamide A β-D-Glucopyranoside from the Seed Kernels of Entadarheedei.Chem Pharm Bull 59: 466-471.
- Antonov AS, Kalinovsky AI, Dmitrenok PS, Kalinin VI, Stonik VA, et al. (2011) New triterpene oligoglycosides from the Caribbean sponge Erylusformosus. Carbohydr Res 346: 2182-2192.
- Yuan WH, Yi YH, Tan RX, Wang ZL, Sun GQ, et al. (2009) Antifungal Triterpene Glycosides from the Sea cucumber Holothuria (Microthele) axiloga. Planta Med75: 647-653.
- Han H, Yi Y, Xu Q, La M, Zhang H (2009) Two New cytotoxic Triterpene Glycosides from the Sea cucumber Holothuriascabra. Planta Med 75: 1608-1612.
- Inoue M, Ohtani K, Kasai R, Okukubo M, Andriantssiferana M, et al. (2009)Cytotoxic 16β-[(D-xylopyranosyl)oxy]oxohexadecanoyl triterpene glycosides from a Malagasy plant, Physenasessiliflora. Phytochem70: 1195-1202.
- Okazaki S, Kinoshita K, Ito S, Koyama K, Yuasa H, et al. (2011) Triterpenoid saponins from Echinopsismacrogona(Cactaceae).Phytochem72: 136-146.
- Gao G, Lu Z, Tao S, Zhang S, Wang F (2011) Triterpenoid saponins with antifeedant activities from stem bark of Catunaregamspinosa(Rubiaceae) against Plutellaxylostella (Plutellidae). Carbohydr Res346: 2200-2205.
- Liu R, Ma SG, Liu YX, Yu SS, Chen XG, et al. (2010) Albizosides D and E, two new cytotoxic triterpene saponins from Albiziachinensis. Carbohydr Res345: 1877-1881.
- Yeom H, Suh JH, Youm JR, Han SB (2012) Simultaneous Determination of Triterpenoid Saponins from Pulsatillakoreana using High Performance Liquid Chromatography coupled with a Charged Aerosol Detector (HPLC-CAD). Bull Korean Chem31: 1159-1164.
- Yamunadevi M, Wesely EG, Johnsom M (2012) Chromatographic finger print studies on saponins of Aervalanata (L.) Juss. Ex. Schultes by using HPTLC. Inter J Curr Pharma Res4: 52-57.
- Theunis MHBL, Foubert K, Pollier J, Gonzalez-Guzman M, Goossens A, et al. (2007) Determination of saponins in Maesalanceolata by LC-UV: Development and validation. Phytochem 68: 2825-2830.
- Wu X, Liu L, Zhang M, Wu D, Wang Y, et al. (2010) Simultaneous analysis of isomers of escinsaponins in human plasma by liquid chromatography-tandem mass spectrometry: Application to a pharmacokinetic study after oral administration. J Chromatog B878: 861-867.
- Song M, Zhang S, Xu X, Hang T, Jia L (2010) Simultaneous determination of three Panaxnotoginsengsaponins at sub-nanograms by LC-MS/MS in dog plasma for pharmacokinetics of compound Danshentablets. J Chromatog B878: 3331-3337.
- Wu W, Qin Q, Guo Y, Sun J, Liu S (2012) Studies on the Chemical Transformation of 20(S)-protopanaxatriol (PPT)-Type Ginsenosides Re, Rg2, and Rf using Rapid Resolution Liquid Chromatography coupled with Quadruple-Time-of-Flight Mass Spectrometry (RRLC-Q-TOF-MS). J Agric Food Chem60: 10007-10014.
- Gomez-Caravaca AM, Segura-Cerretero A, Fernandez A, Caboni MF (2011) Simultaneous Determination of phenolic compounds and saponins in Quinoa (chenopodium quinoaWilld) by a Liquid Chromatography-Diode Array Detection-Electrospray Ionization-Time-of-Flight Mass Spectrometry Methodology. J Agric Food Chem59: 100815-100824.
- Gulcemal D, Masullo M, Napolitano A, Karayildrum T, Bedir E, et al. (2013) Oleanane glycosides from Astragalustauricolus : Isolation and structural elucidation based on a preliminary liquid chromatography-electrospray ionization tandem mass spectrometry profiling. Phytomedicine86: 184-194.
- Gao H, Wang Z, Yao ZH, Wu N, Dong HJ, et al. (2008) Unusual Nortriterpenoid Sponins from Stauntoniachinensis. HelvetChim Acta91: 451-459.
- Gao L, Zhang L, Li N, Liu JY, Cai PL, et al. (2011) New triterpenoid saponins from Patriniascabiosaefolia. Carbohydr Res346: 2881-2885.
- Kakuta K, Baba M, Ito S, Kinoshita K,Koyama K, et al. (2012) New triterpenoid saponins from cacti and antitype I allergy activity of saponins from cactus.Biorg and Med Chem Lett 22: 4793-4800.
- Geyter ED, Lambert E, Geelen D, Smaggha G (2007) Novel Advances with Plant Saponins as Natural Insectides to control Pest Insects. Pest Technology1: 96-105.
- Santo RD (2010) Natural Products as antifungal agents against clinically relevant pathogens. Nat Prod Rep 27: 1084-1098.
- Yuan WH, Yi YH, Tang HF, Liu BS, Wang ZL, et al. (2009) Antifungal Triterpene Glycosides from the Sea cucumber Bohadschiamomorata. Planta Med. 75: 168-173.
- Andesegun SA, Coker HAB, Hamann MT (2008) Antifungal Triterpenoid Saponins from Lecaniodiscuscupanioides. ResJPhytochem2: 93-99.
- Escalante A, Gattuso M, Perez P, Zacchino S (2008) Evidence for the Mechanism of Action of the Antifungal Phytolaccoside B Isolated from PhytolaccatetrameraHauman. J Nat Prod71:1720-1725.
- Sun HX, Xie Y, Ye YP (2009) Advances in saponin-based adjuvants. Vaccine 27: 1787-1796.
- Sun J, Hu S, Song X (2007) Adjuvant effects of protopanaxadiol and protopanaxatriol saponins from ginseng roots on the immune responses to ovalbumin in mice Vaccine25: 1114-1120.
- Yang Z, Chen A, Sun H, Ye Y, Fang W (2007) Ginsenoside Rd elicits Th1 and Th2 immune responses to ovalbumin in mice. Vaccine 25: 161-169.
- Zhai L, Li Y, Wang W, Wang Y, Hu S (2011) Effect of oral administration of ginseng stem-and-leaf saponins (GSLS) on the immune responses to Newcastle disease vaccine in chickens. Vaccine 29: 5007-5014.
- Song X, Bao S, Wu L, Hu S (2009) Ginseng stem-leaf saponins (GSLS) and mineral oil act synergistically to enhance the immune responses to vaccination against foot and mouth disease in mice. Vaccine 27: 51-55.
- Rizk SA, El-garf EM, Talaat AA (2011) Enhancing Effect of Ginseng stem-leaf saponins on the Immune Responses in Vaccinated Calves with FMD Bivalent Vaccine. Inter J Virology 7: 167-175.
- Quan FS, Compans RW, Cho YK, Kang SM (2007) Ginseng and Salviae herbs play a role as immune activators and modulate immune responses during influenza virus infection. Vaccine 25: 272-282.
- Tilwari A, Uma Devi P, Prabha B, Shukla NP (2011) Immunomodulatory Effect of Fractions of Saponins from Tribulusterrestris on Non-Specific Immunity Using In Vitro Phagocytosis. Inter J Drug Discovery and Herbal Res1: 202-207.
- Man S, Gao W, Zhang Y, Huang L, Liu C (2012) Chemical study and medical application of saponins as anti-cancer agents. Fitoterapia81: 703-714.
- Thakur M, Melzig MF Fuchs H, Weng A (2011) Chemistry and pharmacology of saponins: special focus on cytotoxic properties. Botanica:Targets and Therapy 1: 19-28.
- Lacaille-Dubois MA, Pegnyemb DE, Placide Note O, Mittaine-Offer AC (2011) A review of acacic acid-type saponins from Leguminosae-Mimosoideae as potent cytoxic and apoptosis inducing agents. Phytochem Rev10: 565-584.
- Sanchez-Medina A, Stevenson PC, Habtemariam S, Pena-Rodriguez LM, Corcoran O, et al. (2009) Triterpenoid saponins from a cytotoxic root extract of Sideroxylanfoetidissimum subsp.Phytochem70: 765-772.
- Xu QM, Shu Z, He WJ, Chen LY, Yang S, et al. (2012) Antitumor activity of Pulsatillachinensis (Bunge) Regel saponinsin human liver tumor 7402 cells in vitro andin vivo.Phytomedicine19: 293-300.
- Weng A, Bachran D, Gorick C, Bachran C, Fuchs H, et al. (2009) A Simple Method for isolation of GypsohilaSaponins for the combined Application of Targeted Toxins and Saponins in Tumor Therapy. Planta Med75: 1421-1422.
- Tundis R, Bonesi M, Deguin B, Loizzo MR, Menichini F, et al. (2009) Cytotoxic activity andinhibitory effect on nitric oxide of triterpene saponins from the roots of Physospermumverticillatum (Waldst& Kit) (Apiaceae). Bioorg and Med Chem17: 4542-4547.
- Ionokova I, Momekov G, Proksch P (2002) Effects of cycloartanesaponins from hairy roots of AstragalusmembranceusBge., on human tumor cell targets. Fitoterapia 81: 447-451.
- Lee IK, Kang KA, Lim CM, Kim KC, Kim HS et al. (2010) Compound K, a Metabolite of Ginseng saponin, Induces Mitochondria-Dependent and Caspase-Dependent Apoptosis via the Generation of Reactive Oxygen Species in Human Colon Cancer Cells. Int J Mol Sci 11: 4916-4931.
- Liu R, Zhang JZ, Liu WC, Kimura Y, Zheng YN (2010) Anti-Obesity effects of protopanaxadiol types of Ginsenosides isolated from the leaves of American ginseng (Panaxquinquefolius L.) in mice fed with a high-fat diet. Fitoterapia81: 1079-1087.
- Wei GQ, Zheng YN, Li W, Liu WC, Lin T, et al. (2012) Structure modification of ginsenoside Rh2 by fatty acid esterification and its detoxification property in antitumor. Bioorg and Med Chem Letts22: 1082-1085.
- Lee J, Ha YW, Choi TW, Kim HJ, Kim SM, et al. (2011) Cellular Uptake of Ginsenosides in Korean White Ginseng and Red Ginseng and their Apoptotic Activities in Human Breast Cancer Cells. Planta Med77: 133-140.
- Cho SH, Chung KS, Choi JH, Kim DH, Lee KT(2009) Compound K, a metabolite of ginseng saponin, induces apoptosis via caspase-8-dependent pathway in HL-60 human leukemia cells. BioMedCentral Cancer9: 449-461.
- Choo MK, Sakurai H, Kim DH, Saiki I (2008) A ginseng saponin metabolite suppresses human necrosis factor –α-promoted metastasis by suppressing nuclear factor-kB signaling in murine colon cancer cells. Oncology Reports19: 595-600.
- Peng L, Zhou Y, Kong DY, Zhang WD (2010) Antitumor Activities of Dammarane Triterpene Saponins from Bacopa monniera. Phytother Res 24: 864-868.
- Januni P, Sivakumari K, Geetha A, Yuvaraj S, Parthasarathy C (2010) Bacoside A downregulates matrix metalloproteinases 2 and 9 in DEN-induced heptacellular carcinoma. Cell Biochem and Funct 28: 164-169.
- Alam K, Parvez N, Yadav S, Molvi K, Hwisa N, et al. (2011) Antimicrobial activity of leaf callus of Bacopa monnieri L. Pharmacia Lettre3: 287-291.
- Sundarram S, Dwivedi P, Pawar S (2011) Antibacterial Activities of Crude Extracts of Chlorophytumborivilianum to Bacterial Pathogens. Res J Med Plant5: 343-347.
- Bhuvaneswari CH, Rao K, Giri A (2011) Evaluation of GymnemasylvestreAntibacterial Activity in Methanol. Recent Res Sci and Tech3: 73-75.
- Khanna VG, Kannabiran K (2008) Antimicrobial activity of saponin fractions of the leaves of Gymnemasylvestre and Eclipta prostrate. World J MicrobBitech24: 2737-2740.
- Hassan SM, Haq AU, Byrd JA, Berhow MA, Cartwright AL, et al. (2010) Haemolytic and antimicrobial activities of saponin-rich extracts fromguar meal. Food chemistry 119: 600-605.
- Sarikahya NB, Kirmizigul S (2010) Antimicrobia l Triterpenoid Glycosides from Cephalariascoparia. J Nat Prod73: 825-830.
- Liu W, Zhang Y, Han L, Wang H, Saito M, et al. (2008) Saponins (Ginsenosides) from stems and leaves of Panax quinquefolium prevented high-fat diet-induced obesity in mice. Phytomedicine15: 1140-1145.
- Liu J, Shimizu K, Yu H,Zhang C, Jin F, et al. (2010) Stereospecificity of hydroxyl group at C-20 in antiproliferative action of ginsenoside Rh2 on prostate cancer cells. Fitoterapia81: 902-905.
- Hamao M, Matsuda H, Nakamura S, Nakashima S, Semura S, et al. (2011) Anti-obesity effects of the methanolic extract and chakasaponins from the flower buds of Camellia sinensis in mice. Bioorg and Med Chem19: 6033-6041.
- Khanna VG, Kannabiran K, Gett G (2009) Leishmanicidal activity of saponins isolated from the leaves of Eclipta prostrate and Gymnemasylvestre. Ind J Pharmacol 41: 32-35.
- Giunchetti RC, Correa-oliveira R, Martins-Filho OA, Teixeira-Cervalho A, Roatt BM, et al. (2007) Immunogenicity of a killed Leishmania vaccine with saponin adjuvant in dogs. Vaccine 25: 7674-7686.
- Liu J, Shiono J, Shimizu K, Yu H, Zhang C, et al. (2009) 20 (R)-Ginsenoside Rh2 not 20(S) is a selective osteoclastgenesis inhibitor without any cytotoxicity.Bioorg and Med Chem Letts19: 3320-3323.
- Kim DY, Park YG, Quan HY, Kim SJ, Jung MS, et al. (2012) Ginsenoside Rd stimulates the differentiation and mineralization of osteoblastic MC3T3 cells by activating AMP-activated protein kinase via the BMP-2 signaling pathway. Fitotherapia83: 215-222.
- Son II, Park YH, Lee SI, Yang HD, Moon HI (2007) Neuroprotective Activity of Triterpenoid Saponins from Platycode radix against Glutamate-induced Toxicity in Primary Cultured Rat Cortical Cells. Molecules12: 1147-1152.
- Li W, Zhang M, Gu J, Meng ZJ, Zhao LC, et al. (2012) Hypoglycemic effect of protopanaxadiol-type ginsenosides and compound K on Type 2 Diabetes mice induced by High-Fat Diet combining with Streptozotocin via suppression of hepatic gluconeogenesis. Fitoterapia 83: 192-198.
- Nhiem NX, Lim HY, Kiem PV, Minh CV, Thu VK, et al. (2011) Oleanane-type triterpene saponins from the bark of Aralia elata and their NF-jB inhibition and PPAR activation signal pathway. Bio and Med Chem Lett21: 6143-6147.
- Lee J, Lim S, Kang SM, Min S, Son K, et al. (2012) Saponin Inhibits Hepatitis C Virus Propagation by Up-regulating Suppressor of Cytokine Signaling 2. PLOS ONE. 7: E39366.
- Zhao YL, Cai GM, Hong X, Shan LM, Xiao XH (2008) Anti-hepatitis B virus activities of triterpenoid saponin compound from PotentillaansrrineL. Phytomedicine15: 253-258.
- Janani P, Sivakumari K, Parthasarathy C (2009) Hepatoprotective activity of bacoside A against N-nitrosodiethylamine-induced liver toxicity in adult rats. Cell BiolToxicol 25: 425-434.
- Singh A, Quraishi MA (2012) Pipali (Piperlongum) and Brahmi (Bacopa monniera) extracts as green corrosion inhibitor for aluminium in NaOH solution. J Chem Pharm Res 4: 322-325.
- Obor IB, Obi-Egbedi NO (2009) Ginseng Root: A new Efficient and Effective Eco-Friendly Corrosion Inhibitor for Aluminium Alloy of Type AA1060 in Hydrochloric acid Solution. Int J Electrochem Sci4: 1277-1288.
- Abiola OK, Otaigbe JOE (2009) The effects of Phyllanthusamarus extract on corrosion andkinetics of corrosion process of aluminium in alkaline solution. Corr Sci51: 2790-2793.
- Nnanna LA,Nwadiuko OC, Ekekwe ND, Ukpabi CF, Udensi SC, et al. (2011) Adsorption and Inhibitive Properties of Leaf Extract of Newbouldialeavis as a Green Inhibitor for Aluminium Alloy in H2SO4. American J Materials Sci 1: 143-148.
- Ji G, Shukla SK, Dwivedi P, Sundaram S, Prakash R (2011) Inhibitive Effects of Argemonemexicana Plant Extract on Acid Corrosion of Mild Steel. IndEng Che Res 50: 11954-11958.
- Selles C, Benali O, Tabti B, Larabi L, Harek Y (2012) Green corrosion inhibitor: inhibitive action of aqueous extract of Anacyclus pyrethrum L. for the corrosion of mild steel in 0.5M H2SO4. J Mater Environ Sci 3: 206-219.
- Measapyodsuk D, Balsevich J, Reed DW, Covello PS (2007) Saponin Biosynthesis in SaponariavaccariacDNAs Encoding β-Amyrin Synthase and a Triterpene Carboxylic Acid Glucosyltransferase1[OA]. Plant Physiology143: 959-969.
- Naoumkina MA, Modolo LV, Hujman DV, Urbanczyk-Wochniak E, Tang Y, et al. (2010) Genomic and CoexpressionAnalyses Predict MultipleGenes Involved in Triterpene Saponin Biosynthesis in Medicagotruncatula. The Plant Cell22: 850-866.
- Carelli M, Biazzi E, Panara F, Tava A, Scaramelli L, et al. (2011) Medicagotruncatula CYP716A12 is a Multifuctional Oxidase Involved in the Biosynthesis of Hemolytic Saponins. The Plant Cell 23: 3070-3081.
- Mylona P, Owatworakit A, Papadopoulou K, Jenner H, Qin B, et al. (2008)Sad3 and Sad4 are required for Saponin Biosynthesis and Root Development in Oat. The Plant Cell 20: 201-212.
- Han H, Xu QZ, Tang HF, Yi YH, Gong W (2010) Triterpene Glycosides from the Sea Cucumber Pentactaquadrangulari. Planta Med 76: 1900-1904.
- Luo H, Sun C, Sun Y, Wu Q, Li Y, et al. (2011) Analysis of transcriptome of Panaxnotoginseng root uncovers putative triterpene saponin-biosynthesis genes and genetic markers. BMC Genomics 12: 1-15.
- Banerjee M, Shrivastava S (2008) An improved protocol for in vitro multiplication of Bacopa monniera (L.). World J. MicrobiolBiotechnol24: 1355-1359.
- Naik PM,Manoher SH, Praveen N, Murthy HN (2010) Effects of sucrose and pH levels on in vitro shoot regeneration from leaf explants of Bacopa monnieri and accumulation of bacoside A in regenerated shoots. Plant Cell Tiss Organ Cult100: 235-239.
- Majumder S, Garai S, Jha S (2011) Genetic transformation of Bacopa monnieri by wildtype strains of Agrobacterium rhizogenes stimulated production of bacopasaponins in transformed calli and plants. Plant Cell Rep30: 941-954.
- Majumder S, Garai S, Jha S (2012) Use of the cryptogenin gene to stimulate the accumulation of bacopasaponins in transgenic Bacopa monniera plants. Plant Cell Rep31:1899-1909
- Haque R, Saha S, Roy A, Bera T (2009) Micropropagation of an medicinal plant Chlorophytumborivilianum shoot crown bud and characterization and analysis of active ingredients of Roots by TLC. Int J Phi Sci1: 250-260.
- Dave S, Tarafdar JC (2011) Stimulatory synthesis of saponin by mycorrhizali fungi in safedmusli (Chlorophytumborivilianum) tubers.Int Res J Agric Sci1: 137-141.
- Amin E, El-Hawary SS, Fathy MM, Mohammed R, Ali Z, et al.(2011)TriterpenoidalSaponins: Bioactive Secondary Metabolites fromZygophyllumcoccineum.Planta Med77: 488-491.
- Antonov AS, Avilov SA, Kalinovsky AI, Anastyuk SD, Dmitrenok PS (2009) Triterpene Glycosides from Antarctic Sea Cucumbers. 2. Structure of Achlioniceosides A1, A2, and A3 from the Sea Cucumber Achlionicewiolaecuspidata (=Rhipidothuriaracowitzai). J Nat Prod72: 33-38.
- Antonov AS, Avilov SA, Kalinovsky AI, Anastyuk SD, Dmitrenok PS, et al. (2008) Triterpene Glycosides from Antarctic Sea cucumbers. 1. Structures of Liouvillosides A1, A2, A3, B1, B2 from Sea cucumber Staurocucumisliouvillei: New Procedure for Separation of Highly Polar Glycoside Fractions and Taxonomic Revision. JNatProd71: 1677-1685.
- Asao Y, Morikawa T, Xie Y, Okamoto M, Hamao M, et al. (2009) Structures of Acetylated Oleanane-Type Triterpene Saponins, Rarasaponins IV, V, and VI, and Anti-hyperlipidemic Constituents from the Pericarps of Sapindusrarak. Chem Pharm Bull57: 198-203.
- Avilov SA, Silchenko AS, Antonov AS, Kalimin VI, Kalinovsky AI, et al. (2008)Synaptosides A and A1, Triterpene Glycosides from the Sea Cucumber Synapta maculate Containing 3-O-Methylglucuronic Acid and Their Cytotoxic Activity against Tumor cells. JNatProd71: 525-531.
- Bankefors J, Nord LI, Kenne L (2010) Multidimensional profiling of components in complex mixtures of natural products for metabolic analysis, proof of concept: Application of Quillazasaponins. JChromatog B878: 471-476
- Cao S, Brodie PJ, Callmander M, Randrianaivo R, Rakotobe E, et al. (2010)Saponins and a lignan derivative of Terminalia tropophylla from the Madagascar Dry Forest. Phytochem71: 95-99.
- Cao S, Brodie P, Callmander M, Randrianaivo R, RazaÃÆïÃâìÃâÃÂtsalama J, et al. (2009) Antiproliferative Triterpenoid Saponinsof Dodonaeawiscosa from the Madagascar Dry Forest. J Nat Prod 72: 1705-1707.
- Chen Q, Luo JG, Kong LY (2011) New triterpenoid saponins from the roots of Gypsophila perfoliata Linn. Carbohydr Res346: 2206-2212.
- Choi YH, Yoo DS, Cha MR, Choi CW, Kim YS, et al. (2010) Antiproliferative Effects of Saponins from the Roots of Platycodon grandiÃÆïÃâìÃâââ¬Å¡orum on Cultured Human Tumor Cells. J Nat Prod 73: 1863-1867.
- CiofÃÆïÃâìÃâàG, Dal Piaz F, Vassallo A, Venturella F, De Caprariis P, et al. (2009) Antiproliferative Oleanane Saponins from Merytadenhamii. J Nat Prod 71: 1000-1004.
- Dall’Acqua S, Castagliuolo I, Brun P, Ditadi F, Palù P, et al. (2010) Triterpene glycosides with in vitro anti-inÃÆïÃâìÃâââ¬Å¡ammatory activity from Cyclamen repandum tubers. Carbohydr Res345: 709-714.
- Fan L, Lu J, Xu B, Gao S, Zhang H, et al. (2010) Oleanane Saponins from Rhizome of Anemone raddeana. HelvetChim Acta93: 58-64.
- Feng YL, Wu B, Li HR, Li YQ, Xu LZ, et al. (2009)TriterpenoidalSaponins from the barks of Zygophyllumfabago L. Chem Pharm Bull 56: 858-860.
- Fu Q, Zan K, Zhao M, Zhou S, Shi S, et al. (2010) Triterpene Saponins from Clematis chinensis and Their Potential Anti-inÃÆïÃâìÃâââ¬Å¡ammatory Activity. J Nat Prod73: 1234-1239.
- Fu X, Li XC, Smillie TI, Carvalho P, Mabusela WS, et al. (2008)Cycloartane Glycosides from Sutherlandiafrutescens. JNatProd 71: 1749-1753.
- Fu HZ, Li CJ, Yang JZ, Shen ZF, Zhang DM (2011) Potential Anti-inflammatory Constituents of the Stems of Gordoniachrysandra. JNatProd74: 1066-1072.
- Gan M, Liu M, Gan L, Lin S, Liu B, et al. (2012) Dammarane Glycosides from the Root of Machilusyaoshansis. J Nat Prod75: 1373-1382.
- Garai S, Garai S (2010) Corrosion inhibition of mild steel in acid solution by bacopasaponins from Bacopa monniera.The International Symposium on Current Status and Opportunities in Aromatic and Medicinal Plants.
- Gao H, Zhao F, Chen GD, Chen SD, Yu Y, et al. (2009)Bidesmoside triterpenoid glycosides from Stauntoniachinensis and relationship to anti-inÃÆïÃâìÃâââ¬Å¡ammation. Phytochem 70: 795-806.
- Han JY, In JG, Kwon YS, Choi YE (2010) Regulation of ginsenoside and phytosterol biosynthesis by DNA interferences of squalene epoxidase gene in Panax ginseng. Phytochem 71: 36-46.
- Huang HC, Liaw CC, Zhang LJ, Ho HU, Yang Kuo LM, et al. (2008)Triterpenoidalsaponins from Hydrocotylesibthorpioides.Phytochem 69: 1597-1603.
- Huang X, Liang Y, Cai X, Feng X, Zhang C, et al. (2009) Four New cytotoxic oligosaccharidic derivatives of 12-oleanene from LysimachiaheterogeneaKlatt. Biorg. & Med. Chem. Letts 19: 6515-6518.
- Lee JH, Jeon J, Lee YJ, Lee HS, Sim CJ, et al. (2012)Nortriterpene Glycosides of the Sarasinoside Class from the Sponge Lipastrotethya sp. J. Nat. Prod. 75: 1365-1372.
- Li ZJ, Chen JC, Chen Y, Sun Y, Song NL, et al. (2009) Three New Triterpene Saponins from Hemsleyachinensis. Helvet. Chim Acta 92: 1853-1859.
- Li W, Bi X, Wang K, Li D, Satou T, et al. (2009) Triterpenoid saponins from Impatiens siculifer. Phytochem 70: 816-821.
- Li C, Yang J, Yu S, Chen N, Xue W, et al. (2008) Triterpenoid Saponins with Neuroprotective Effects from the Roots of Polygala tenuifolia. Planta Med 74: 133-141.
- Li W, Fu H, Bai H, Sasaki T, Kato H, et al. (2009) Triterpenoid Saponins from Rubusellipticusvar. Obcordatus. J Nat Prod72: 1755-1760.
- Li F, Li W, Fu H, Zhang H, Koike K (2007) Pancreatic Lipase-Inhibiting Triterpenoid Saponins from Fruits of Acanthopanaxsenticosus. Chem Pharm Bull55: 1087-1089.
- Liang D, Hao GY, Zhang GJ, Zhang QJ, Chen RY, et al. (2011) Cytotoxic Triterpenoid Saponins from Lysimachiaclethroides. J Nat Prod74: 2128-2136.
- Liu R, Ma S, Yu S, Pei Y, Zhang S, et al. (2009) Cytotoxic Oleanane Triterpene Saponins from Albiziachinensis. J Nat Prod72: 632-639.
- Litaudon M, Jolly C, Callonec CL, Cuong DD, Retailleau P, et al. (2009) Cytotoxic Pentacyclic Triterpenoids from Combretum sundaicum and Lantana camara as Inhibitors of Bcl-xL/BakBH3 Domain Peptide Interaction. J Nat Prod72: 1314-1320.
- Ma L, Gu YC, Luo JG, Wang JS, Huang XF, et al. (2009) Triterpenoid Saponins from Dianthus versicolor. J Nat Prod72: 640-644.
- Ma J, Chen Q, Lai D, Sun W, Zhang T, et al. (2010) Separation and purification of triterpene saponins from roots of Radix phytolaccae by high-speed countercurrent chromatography coupled with evaporative light scattering detection.J. chromatogandRel Techno33: 563-571.
- Ma SG, Hu YC, Yu SS, Zhang Y, Chen XG, et al. (2008) Cytotoxic Triterpenoid Saponins Acylated with Monoterpenic Acid from Pithecellobium lucidum. J Nat Prod71: 41-46.
- Magid AA, Bobichon H, Borie N, Lalun N, Long C, et al. (2010) Cytotoxic triterpenoid saponins from the stem bark of Antonia ovate. Phytochem 71: 429-434.
- MagidAA, Lalun N, Long C, Borie N, Bobichon H, et al. (2012) Triterpene saponins from Antonia ovata leaves. Phytochem 77: 268-274.
- Mazzola EP, Perkinson A, Kennelly EJ, Coxon B, Einbond LS, et al. (2011) Utility of coupled HSQC experiments in the intact structural elucidation of three complex saponins from Blighiasapida.Carbohydr Res 346: 759-768.
- Mehta BK, Mehta P, Gupta M (2009) A new naturally acetylated triterpene saponin from Nigella sativa. Carbohydr Res344: 149-151.
- Melek FR, Miyase T, Ghaly TN, Nabil M (2007) Triterpenoid saponins with N-acetyl sugar from the bark of Albiziaprocera. Phytochem 68: 1261-1266.
- Mencherini T, Picerno P, Del Gaudio P, Festa M, Capasso A, et al. (2009)Saponins and Polyphenols from Fadogiaancylantha(MakoniTea). J Nat Prod73: 247-251.
- Meng D, Qiang S, Lou L, Zhao W (2009) Cytotoxic Cucurbitane-Type Triterpenoids from Elaeocarpushainanensis. Planta Med74: 1741-1744.
- Miyase T, Melek FR, Ghaly NS, Warashina T, El-Kady M, et al. (2010)Echinocystic acid, 3, 16-O-bisglycosides from Albiziaprocera. Phytochem 71: 1375-1380.
- Miyase T, Melek FR, Warashina T, Selim MA, El Fiki NM, et al. (2010) Cytotoxic triterpenoid saponins acylated with monoterpenic acids from fruits of GleditsiacaspicaDesf.Phytochem 71: 1908-1916.
- Morikawa T, Matsuda H, Li N, Li X, Yoshikawa M (2007) Bioactive Saponins and Glycosides Part 291) Acylated Oleanane-Type Triterpene Saponins: Theasaponins A6, A7, and B5 from the Seeds of Camellia sinensis. Helvet. Chim Acta 90: 2342-2347.
- Morikawa T, Xie Y, Asao Y, Okamoto M, Yamashita C, et al. (2009) Oleanane-type triterpene oligoglycosides with pancreatic lipase inhibitory activity from the pericarps of Sapindusrarak. Phytochem 70: 1166-1172.
- Morikawa T, Li X, Nishida E, Ito Y, Matsuda H, et al. (2008)Perennisosides I-VII, Acylated Triterpene Saponins with Antihyperlipidemic Activities from the Flowers of Bellis perenni. J Nat Prod71: 828-835.
- Mu LH, Wei NY, Liu P (2012) Cytotoxic Triterpenoid Saponins from Ardisiagigantifolia. Planta Med.78: 617-621.
- Nhiem NX, Kiem PV, Minh CV, Ha DT, Tai DBH, et al. (2010)Lupane Triterpene Glycosides from Leave of Acanthopanaxkoreanum and Their in vitro Cytotoxicity.Planta Med 76: 189-194.
- Nhiem NX, Kiem PV, Minh CV, Tai BH, Tung NH, et al. (2010) Structure–activity relationship of lupane-triterpene glycosides from Acanthopanaxkoreanum on spleen lymphocyte IL-2 and IFN-γ.Bio and MedChem Lett20: 4927-4931.
- Placide Note O,Mitaine-Offer AC, Miyamoto T, Paululat T, Mirjolet JF, et al. (2009) Cytotoxic Acacic Acid Glycosides from the Roots of Albiziacoriaria. J Nat Prod72: 1725-1730.
- Perrone A, Masullo M, Bassarello C, Bloise E, Hamed A, et al. (2008) Unusual cycloartane glycosides from Astragaluseremophilus. Tetrahedron 64: 5061-5071.
- Ponou BK, Teponno RB, Ricciutelli M, Quassinti L, Bramucci M, et al. (2010) Dimeric antioxidant and cytotoxic triterpenoid saponins from Terminalia ivorensis A. Chev.Phytochem 71: 2108-2115.
- Quang TH, Ngan NTT, Minh CV, Kiem PV, Nhiem NX, et al. (2011) Anti-inflammatory Triterpenoid Saponins from the Stem Bark of Kalopanaxpictus. JNatProd74: 1908-1915.
- Quang TH, Ngan NTT, Minh CV, Kiem PV, Thao NP, et al. (2011) Effect of triterpene saponins from the stem bark of Kalopanaxpictus on the transactivational activities of three PPAR subtypes. Carbohydr Res346: 2567-2575.
- Sánchez-Medina A, Stevenson PC, Habtemariam S, Peña-Rodríguez LM, Corcoran O, et al. (2009) Triterpenoid saponins from a cytotoxic root extract of Sideroxylonfoetidissimum subsp. Gaumeri. Phytochem 70: 765-772.
- Scognamiglio M, Abrosca BD, Fiumano V, Chambery A, Severino V, et al. (2012) Oleanane saponins from Bellis sylvestris Cyr. and evaluation of their phytotoxicity on Aegelopsgeniculata Roth. Phytochem84: 125-134.
- Silchenko AS, Avilov SA, Kalinin VI, Kalinovsky AI, Dmitrenok PS, et al. (2008) Constituents of the Sea Cucumber Cucumariaokhotensis. Structures of Okhotosides B1–B3 and Cytotoxic Activities of Some Glycosides from this Species.J Nat Prod71: 351-356.
- Sumathi T, Nongbri A (2008) Hepatoprotective effect of Bacoside A, a major constituentBacopa monnieraLinn. Phytomedicine15: 901-905.
- Sumathi T, Devaraj SN (2009) Effect of Bacopa monnieraon liver and kidney toxicity inchronic use of opioids. Phytomedicine16: 897-903.
- Tabopda TK, Mitaine-Offer AC, Miyamoto T, Tanaka C, Mirjolet JF, et al. (2012) Triterpenoid saponins from Hydrocotylebonariensis. Lam Phytochem73: 142-147.
- Tabopda TK, Ngoupayo J, Khan Tanoli SA, Mitaine-Offer AC, Ngadjui BT et al. (2009) Antimicrobial Pentacyclic Triterpenoidsfrom Terminalia superb. Planta Med75: 522-527.
- Tapondjou LA, Nyaa LBT, Tane P, Ricciutelli M, Quassinti L, et al. (2011) Cytotoxic and antioxidant triterpene saponins from Butyrospermumparkii (Sapotaceae). Carbohydr Res346: 2699-2704.
- Tava A, Mella M, Avato P, Biazzi E, Pecetti L, et al. (2009) New Triterpene Saponins from the Aerial Parts ofMedicagoarabica (L.) Huds. J. Agric Food Chem57: 2826-2835.
- Tian Y, Tang HF, Qiu F, Wang XJ, Chen XL,et al.(2009) Triterpenoid Saponins fromArdisiapusilla and their Cytotoxic Activity. Planta Med75: 70-75.
- Timité G, Mitaine-Offer AC, Miyamoto T, Tanaka C, Mirjolet JF, et al.(2011) Unusual oleanane-type saponins from Arenariamontana. Phytochem 72: 503-507.
- Timite G, Mitaine-Offer AC, Miyamoto T, Ramezani M, Rustaiyan A, et al. (2010) Structure elucidation of new oleanane-type glycosides from three species of Acanthophyllum. MagResChem48: 370-374.
- Ukiya M, Akihisa T, Yasukawa K, Koike K, Takahashi A, et al. (2007) Triterpene Glycosides from the Flower Petals of Sunflower (Helianthus annuus) and Their Anti-inflammatory Activity.J Nat Prod 70: 813-816.
- Wang Y, Zhang D, Ye W, Yin Z, Fung KP, et al. (2008) TriterpenoidSaponins from Androsaceumbellata and their Anti-proliferative Activities in Human Hepatoma Cells. Planta Med 74: 1280-1284.
- Wang XY, Chen XL, Tang HF, Gao H, Tian XR, et al.(2011) Cytotoxic Triterpenoid Saponins from the Rhizomes of Anemone taipaiensis. Planta Med 77: 1550-1554.
- Wang H, Gao J, Kou J, Zhu D, Yu B (2008)Anti-inflammatory activities of triterpenoid saponins from Polygala japonica. Phytomedicine15: 321-326.
- Wang L, Cai Y, Zhang XQ, Fan CL, Zhang QW, et al.(2012) New triterpenoid glycosides from the roots of Ilex asprella. Carbohydr Res 349: 39-43.
- Xu WH, Jacob MR, Agarwal AK, Clark AM, Liang ZS, et al. (2009) VerbesinosidesA-F,15,27-Cyclooleanane Saponins from the American Native Plant Verbesinavirginica.J Nat Prod72: 1022-1027.
- Xu LP, Wang H, Yuan Z (2008) Triterpenoid Saponins with Anti inflammatory Activity fromCodonopsislanceolata. Planta Med74: 1412-1415.
- Yang H, Jeong EJ, Kim J, Sung SH, Kim YC (2011) Antiproliferative Triterpenes from he Leaves and Twigs of Juglans sinensis on HSC-T6 Cells. JNatProd74: 751-756.
- Yang H, Cho HJ, Sim SH, Chung YK, Kim DD, et al. (2012) Cytotoxic terpenoids from Juglans sinensis leaves and twigs. Bio and MedChem Letts22: 2079-2083.
- Yokosuka A, Sano T, Hashmoto K, Sakagami H, Mimaki Y (2009) Triterpene Glycosides from the Whole Plant of Anemone hupehensisvar. japonica and Their Cytotoxic Activity. Chem Pharm Bull 57: 1425-1430.
- Yoon JH, Choi YJ, Cha SW, Lee SG (2012) Anti-metastatic effects of ginsenoside Rd via inactivation of MAPK signaling and induction of focal adhesion formation. Phytomedicine19: 284-292.
- Yoshikawa M, Li X, Nishida E, Nakamura S, Matsuda H, et al. (2008) Medicinal Flowers. Structures of Perennisaponins A, B, C, D, E and F, Acylated Oleanane-Type Triterpene Oligoglycosides, from the Flowers of Bellis perennis. Chem Pharm Bull56 559-568.
- Yu L, Yang JZ, Chen XG, Shi JG, Zhang DM (2009) Cytotoxic Triterpenoid Glycosides from the Roots ofGordoniachrysandra. J Nat Prod74:866-870.
- Yu L, Wang X, Wei X, Wang M, Chen L, et al. (2010) Triterpenoid saponins from Xanthocerassorbifolia Bunge and their inhibitory activity on human cancer cell lines. Bioorgand Med Chem Letts22: 5232-5238.
- Yuan WH, Yi YH, Tang HF, Liu BS, Wang ZL, et al. (2009) Antifungal Triterpene Glycosides from the Sea Cucumber Bohadschiamarmorata. Planta Med75 168-173.
- Yuan W, Yi Y, Tang H, Xue M, Wang Z, et al. (2008) Two New Holostan-Type Triterpene Glycosides from the Sea Cucumber Bohadschiamarmorata JAEGER. Chem Pharm Bull56: 1207-1211.
- Yuan W, Wang P, Deng G, Li S (2012) Cytotoxic triterpenoid saponins from AesculusglabraWilld. Phytochem 75: 67-77.
- Zhang Z, Li S (2007) Cytotoxic triterpenoid saponins from the fruits of Aesculuspavia L. Phytochem 68: 2075-2086.
- Zhang H, Samadi AK, Rao KV, Cohen MS, Timmermann BN (2011) Cytotoxic Oleanane-type Saponins from Albiziainundata. J Nat Prod74: 477-482.
- Zheng Q, Li W, Han L, Koike K (2007) Pancreatic Lipase-Inhibiting Triterpenoid Saponins from Gypsophila oldhamiana. Chem Pharm Bull55:646-650.
- Zhou M, Xu M, Ma XX, Zheng K, Yang K, et al. (2012) Antiviral Triterpenoid Saponins from the Roots of Ilex asprella. Planta Med78: 1702-1705.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences